
Acta Horticulturae Sinica ›› 2023, Vol. 50 ›› Issue (3): 534-548.doi: 10.16420/j.issn.0513-353x.2022-0003
• Research Papers • Previous Articles Next Articles
MAO Kexin1,2, AN Miao1, WANG Hairong1, WANG Shijin3, LÜ Wei2, GUO Yingtian1, LI Jian1,*( ), LI Guotian1,*(
), LI Guotian1,*( )
)
Received:2022-09-03
Revised:2022-12-30
Online:2023-03-25
Published:2023-04-03
Contact:
*(E-mail:lijian097597@163.com,ligt2008@163.com)
CLC Number:
MAO Kexin, AN Miao, WANG Hairong, WANG Shijin, LÜ Wei, GUO Yingtian, LI Jian, LI Guotian. Identification and Low Temperature Expression Analysis of MYB Transcription Factor Family in Kiwifruit[J]. Acta Horticulturae Sinica, 2023, 50(3): 534-548.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2022-0003
| 基因Gene | 上游引物(5′-3′)Forward primer | 下游引物(5′-3′)Reverse primer |
|---|---|---|
| AcMYB200 | GAGATACCAGTGGCGGAGAG | CCCTGTATGCCGTTGCTTAT |
| AcMYB61 | TCTTGGATGGTGTTGTTCCA | CCTGATCTGCTACGCTTTCC |
| AcMYB63 | GAGGTGCTTAAGCCAACAGC | GTCCGACTTCCAAACTGCAT |
| AcMYB56 | GTTCATAAGAGCGGGACCAA | GGAAGCATTTGGGTCAAAAA |
| AcMYB71 | GAACCGGTGGTATTGAAACG | ACACGATCTCCACCTTTTGG |
| AcMYB123 | CGACAAGTTCCTCGAAGCTC | TTTTGCGGGTAAGGATGAAC |
| AcMYB210 | TGTGAATCCTCCACAAGCTG | ATGTGTCCCCGTTTTAGCAG |
| AcMYB235 | CGACCTGAGCAAGAAGATCC | CTTCTGTGCATGGCTTCGTA |
| Actin | GTGCTCAGTGGTGGTTCAA | GACGCTGTATTTCCTCTCAG |
Table 1 Primers in this study
| 基因Gene | 上游引物(5′-3′)Forward primer | 下游引物(5′-3′)Reverse primer |
|---|---|---|
| AcMYB200 | GAGATACCAGTGGCGGAGAG | CCCTGTATGCCGTTGCTTAT |
| AcMYB61 | TCTTGGATGGTGTTGTTCCA | CCTGATCTGCTACGCTTTCC |
| AcMYB63 | GAGGTGCTTAAGCCAACAGC | GTCCGACTTCCAAACTGCAT |
| AcMYB56 | GTTCATAAGAGCGGGACCAA | GGAAGCATTTGGGTCAAAAA |
| AcMYB71 | GAACCGGTGGTATTGAAACG | ACACGATCTCCACCTTTTGG |
| AcMYB123 | CGACAAGTTCCTCGAAGCTC | TTTTGCGGGTAAGGATGAAC |
| AcMYB210 | TGTGAATCCTCCACAAGCTG | ATGTGTCCCCGTTTTAGCAG |
| AcMYB235 | CGACCTGAGCAAGAAGATCC | CTTCTGTGCATGGCTTCGTA |
| Actin | GTGCTCAGTGGTGGTTCAA | GACGCTGTATTTCCTCTCAG |
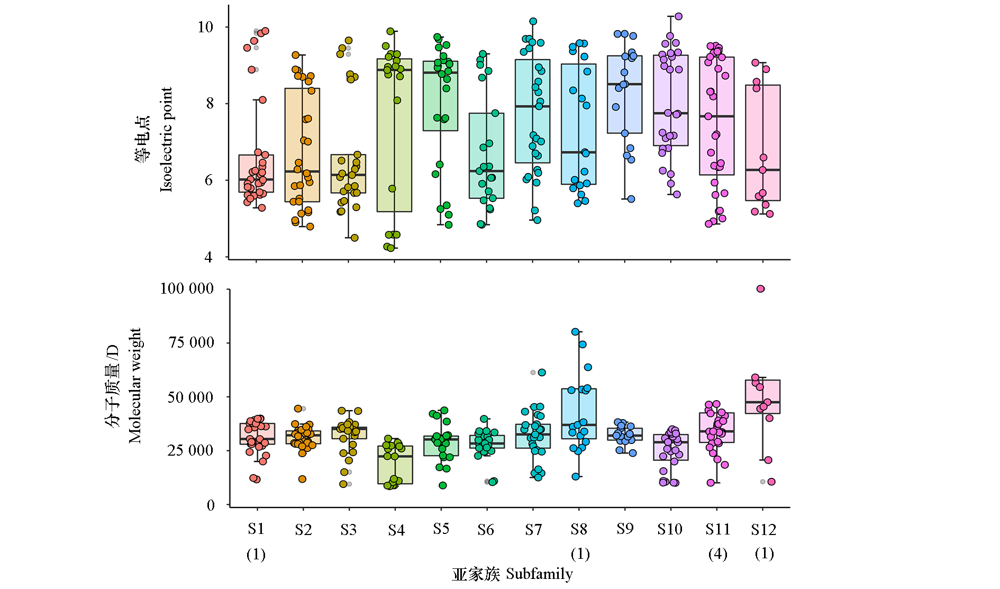
Fig. 2 Protein isoelectric point and molecular weight of kiwifruit MYB family The number in parenthes indicates the number of MYB proteins which molecular weight exceeding 100 000 D.
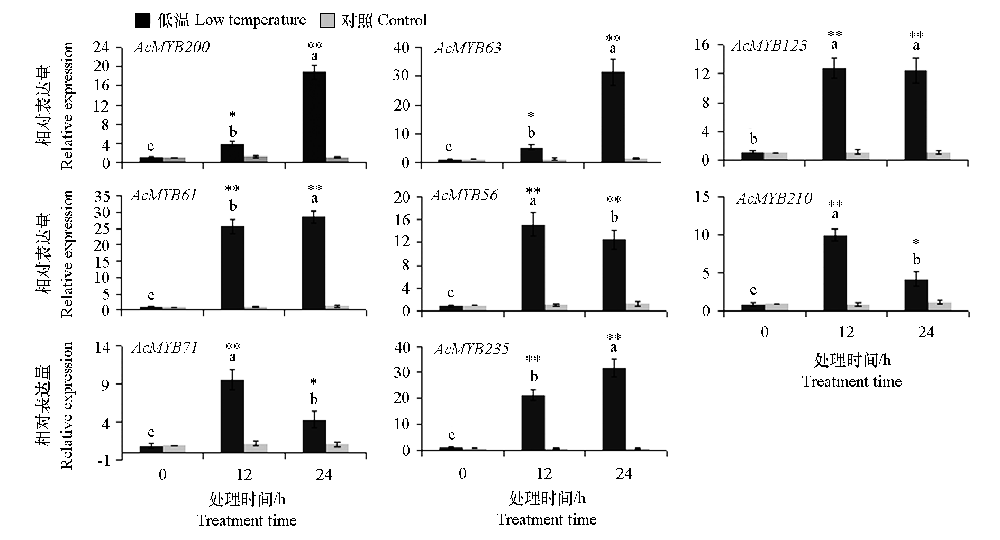
Fig. 8 Expression analysis of eight kiwifruit MYB genes under low temperature treatment Different letters indicate that there are significant differences in genes at the level of 0.05 under different treatment times. *,** indicated significant difference between treatment and control at 0.05 and 0.01 levels,respectively.
| [1] |
Aasland R, Stewart A F, Gibson T. 1996. The SANT domain:a putative DNA-binding domain in the SWI-SNF and ADA complexes,the transcriptional co-repressor N-CoR and TFIIIB. Trends in Biochemical Sciences, 21:87-88.
pmid: 8882580 |
| [2] |
Anwar M, Yu W, Yao H, Zhou P, Allan A C, Zeng L. 2019. NtMYB3,an R2R3-MYB from narcissus,regulates flavonoid biosynthesis. International Journal of Molecular Sciences, 20:5456.
doi: 10.3390/ijms20215456 URL |
| [3] | Ashesh N. 2002. Investigations on evolutionary changes in base distributions in gene sequences. Internet Electronic Journal of Molecular Design, 1 (10):545. |
| [4] |
Baldoni E, Genga A, Cominelli E. 2015. Plant MYB transcription factors:their role in drought response mechanisms. International Journal of Molecular Sciences, 16:15811-15851.
doi: 10.3390/ijms160715811 pmid: 26184177 |
| [5] |
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden T L. 2009. BLAST+:architecture and applications. BMC Bioinformatics, 10 (421):1-9.
doi: 10.1186/1471-2105-10-1 |
| [6] |
Cao Y, Li K, Li Y, Zhao X, Wang L. 2020. MYB transcription factors as regulators of secondary metabolism in plants. Biology, 9 (3):61-76.
doi: 10.3390/biology9030061 URL |
| [7] |
Chen C, Chen H, Zhang Y, Thomas H R, Frank M H, He Y, Xia R. 2020. TBtools:an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 13 (8):1194-1202.
doi: 10.1016/j.molp.2020.06.009 URL |
| [8] |
Chen Y H, Wu X M, Ling H Q, Yang W C. 2006a. Transgenic expression of DwMYB2 impairs iron transport from root to shoot in Arabidopsis thaliana. Cell Research, 16:830-840.
doi: 10.1038/sj.cr.7310099 |
| [9] |
Chen Y H, Yang X Y, He K, Liu M H, Li J G, Luo J C, Deng X W, Chen Z L, Gu H Y, Qu L J. 2006b. The MYB transcription factor superfamily of Arabidopsis:expression analysis and phylogenetic comparison with the rice MYB family. Plant Molecular Biology, 60:107-124.
doi: 10.1007/s11103-005-2910-y URL |
| [10] | Das G, Duncan L R. 2014. Configuration of wobble base pairs having pyrimidines as anticodon wobble bases: significance for codon degeneracy. Journal of Biomolecular Structure and Dynamics, 329:1500-1520. |
| [11] |
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. 2010. MYB transcription factors in Arabidopsis. Trends in Plant Science, 15:573-581.
doi: 10.1016/j.tplants.2010.06.005 URL |
| [12] |
Enany S. 2014. Structural and functional analysis of hypothetical and conserved proteins of Clostridium tetani. Journal of Infection and Public Health, 7 (4):296-307.
doi: 10.1016/j.jiph.2014.02.002 URL |
| [13] |
Feng G, Burleigh J G, Braun E L, Mei W, Barbazuk W B. 2017. Evolution of the 3R-MYB gene family in plants. Genome Biology and Evolution, 9:1013-1029.
doi: 10.1093/gbe/evx056 pmid: 28444194 |
| [14] | Gupta S K, Majumdar S K, Bhattacharya T. 2000. Studies on the relationships between the synonymous codon usage and protein secondary structural units. Biochemical & Biophysical Research Communications, 269 (3):692-696. |
| [15] |
Haupt S, Ziegler A, Cowan G, Torrance L. 2009. Studies of the role and function of barley stripe mosaic virus encoded proteins in replication and movement using GFP fusions. Methods in Molecular Biology, 515:287-297.
doi: 10.1007/978-1-59745-559-6_20 pmid: 19378124 |
| [16] | Hou Qian-qian. 2019. Expression strategy and application of exogenous genes in Candida glycerinogenes[Ph. D. Dissertation]. Wuxi: Jiangnan University. (in Chinese) |
| 侯倩倩. 2019. 外源基因在产甘油假丝酵母中的表达策略及应用研究[博士论文]. 无锡: 江南大学. | |
| [17] | Huang S X, Ding J, Deng D J, Tang W, Sun H H, Xiao F M, Wang H L, Zheng H K, Fei Z J, Liu Y S. 2013. Draft genome of the kiwifruit Actinidia chinensis. Nature Communications, 4 (4):26-40. |
| [18] | Hughes A L. 1994. The evolution of functionally novel proteins after gene duplication. Proceedings of the Royal Society B-biological Sciences, 256:119-124. |
| [19] |
Jia J, Xue Q Z. 2009. Codon usage biases of transposable elements and host nuclear genes in Arabidopsis thaliana and Oryza sativa. Genomics Proteomics Bioinformatics, 7 (4):175-184.
doi: 10.1016/S1672-0229(08)60047-9 URL |
| [20] |
Jiang C K, Rao G Y. 2020. Insights into the diversification and evolution of R2R3-MYB transcription factors in plants. Plant Physiology, 183:637-655.
doi: 10.1104/pp.19.01082 URL |
| [21] |
Kanei-Ishii C, Sarai A, Sawazaki T, Nakagoshi H, He D N. 1990. The tryptophan cluster:a hypothetical structure of the DNA-binding domain of the MYB protooncogene product. Journal of Biological Chemistry, 265:19990-19995.
pmid: 2246275 |
| [22] | Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y. 2002. PlantCARE,a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research, 30:325-327. |
| [23] |
Letunic I, Khedkar S, Bork P. 2020. SMART:recent updates,new developments and status in 2020. Nucleic Acids Research, 49:458-460.
doi: 10.1093/nar/gkaa1189 URL |
| [24] |
Li Guisheng. 2021. Comparative analysis of‘Jinyan’and‘Hongyang’kiwifruit transcriptomes. Acta Horticulturae Sinica, 48 (6):1183-1196. (in Chinese)
doi: 10.16420/j.issn.0513-353x.2020-0510 |
|
李贵生. 2021. 猕猴桃‘金艳’和‘红阳’果实转录组的比较分析. 园艺学报, 48 (6):1183-1196.
doi: 10.16420/j.issn.0513-353x.2020-0510 |
|
| [25] | Li J, Han G, Sun C, Sui N. 2019. Research advances of MYB transcription factors in plant stress resistance and breeding. Plant Signaling & Behavior, 14 (8):e1613131. |
| [26] |
Li W, Ding Z, Ruan M, Yu X, Peng M, Liu Y. 2017. Kiwifruit R2R3-MYB transcription factors and contribution of the novel AcMYB75 to red kiwifruit anthocyanin biosynthesis. Scientific Reports, 7 (1):16861.
doi: 10.1038/s41598-017-16905-1 |
| [27] |
Liu C, Xie T, Chen C, Luan A, Long J, Li C, Ding Y, Yehua H. 2017. Genome-wide organization and expression profiling of the R2R3-MYB transcription factor family in pineapple(Ananas comosus). BMC Genomics, 18:503-518.
doi: 10.1186/s12864-017-3896-y URL |
| [28] |
Liu J, Osbourn A, Ma P. 2015. MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Molecular Plant, 8:689-708.
doi: 10.1016/j.molp.2015.03.012 pmid: 25840349 |
| [29] | Lu S, Wang J, Chitsaz F, Derbyshire M K, Geer R C. 2020. CDD/SPARCLE:the conserved domain database in 2020. Nucleic Acids Research, 48:265-268. |
| [30] | Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar Gustavo A. 2020. Pfam:the protein families database in 2021. Nucleic Acids Research, 49:412-419. |
| [31] |
Novembre J A. 2002. Accounting for background nucleotide composition when measuring codon usage bias. Molecular Biology and Evolution, 19 (8):1390-1394.
pmid: 12140252 |
| [32] |
Potter S C, Luciani A, Eddy S R, Park Y, Lopez R, Finn R D. 2018. HMMER web server:2018 update. Nucleic Acids Research, 46:200-204.
doi: 10.1093/nar/gky448 pmid: 29905871 |
| [33] | Roy S. 2016. Function of MYB domain transcription factors in abiotic stress and epigenetic control of stress response in plant genome. Plant Signaling & Behavior, 11:e1117723. |
| [34] | Shi Yang, Xu Xiao, Li Hao-yang, Xu Qian, Xu Jichen. 2014. Bioinformatics analysis of the expansin gene family in rice. Hereditas, 36 (8):809-820. (in Chinese) |
| 施杨, 徐筱, 李昊阳, 徐倩, 徐吉臣. 2014. 水稻扩展蛋白家族的生物信息学分析. 遗传, 36 (8):809-820. | |
| [35] |
Sueoka N. 2001. Near homogeneity of PR2-bias fingerprints in the human genome and their implications in phylogenetic analyses. Journal of Molecular Evolution, 53 (4-5):469-476.
pmid: 11675607 |
| [36] | Suzuki Y. 2011. Statistical methods for detecting natural selection from genomic data. Genes & Genetic Systems, 856:359-376. |
| [37] |
Tamura K, Stecher G, Kumar S. 2021. MEGA11:molecular evolutionary genetics analysis version 11. Molecular Biology and Evolution, 38:3022-3027.
doi: 10.1093/molbev/msab120 URL |
| [38] | Tian F, Yang D C, Meng Y Q, Jin J, Gao G. 2020. PlantRegMap:charting functional regulatory maps in plants. Nucleic Acids Research, 48:1104-1113. |
| [39] |
Wang L, Roossinck M J. 2006. Comparative analysis of expressed sequences reveals a conserved pattern of optimal codon usage in plants. Plant Molecular Biology, 61:699-710.
doi: 10.1007/s11103-006-0041-8 pmid: 16897485 |
| [40] | Wang L, Tang W, Hu Y, Zhang Y, Sun J. 2019. A MYB/bHLH complex regulates tissue-specific anthocyanin biosynthesis in the inner pericarp of red-centered kiwifruit Actinidia chinensis cv. Hongyang. Plant Journal, 99:359-378. |
| [41] |
Wang X, Niu Y, Zheng Y. 2021. Multiple functions of MYB transcription factors in abiotic stress responses. International Journal of Molecular Science, 22:6125.
doi: 10.3390/ijms22116125 URL |
| [42] |
Wang Y, Tang H, Debarry J D, Tan X, Li J. 2012. MCScanX:a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Research, 40:e49.
doi: 10.1093/nar/gkr1293 URL |
| [43] |
Wilkins M R, Gasteiger E, Bairoch A, Sanchez J C, Williams K L. 1999. Protein identification and analysis tools in the ExPASy server. Methods in Molecular Biology, 112:531-552.
pmid: 10027275 |
| [44] |
Wu H L, Ma T, Kang M H, Ai F D, Zhang J L, Dong G Y, Liu J Q. 2019. A high-quality Actinidia chinensis(kiwifruit)genome. Horticulture Research, 6:117.
doi: 10.1038/s41438-019-0202-y |
| [45] |
Wu Jiacheng, Wang Guiqing, Muhammad Anwar, Zeng Lihui. 2018. Cloning and functional analysis of R2R3-MYB gene NtMYB5 in Narcissus tazetta var. chinensis. Acta Horticulturae Sinica, 45 (7):1327-1337. (in Chinese)
doi: 10.16420/j.issn.0513-353x.2017-0889 |
|
吴嘉诚, 王桂青, Muhammad Anwar, 曾黎辉. 2018. 中国水仙R2R3-MYB 基因NtMYB5的克隆和功能研究. 园艺学报, 45 (7):1327-1337.
doi: 10.16420/j.issn.0513-353x.2017-0889 |
|
| [46] |
Yan H L, Pei X N, Zhang H, Li X, Zhang X X, Zhao M H, Chiang V L, Sederoff R R, Zhao X Y. 2021. MYB-mediated regulation of anthocyanin biosynthesis. International Journal of Molecular Sciences, 22 (6):3103-3128.
doi: 10.3390/ijms22063103 URL |
| [47] |
Yan X W, Hoek T A, Vale R D, Marvin M E. 2016. Dynamics of translation of single mRNA molecules invivo. Cell, 165:976-989.
doi: 10.1016/j.cell.2016.04.034 URL |
| [48] | Yang Ping. 2011. Characteristics analysis of codon usage of rice HKT gene family. Journal of Shanxi Agricultural Sciences, 39 (11):1137-1140. (in Chinese) |
| 杨平. 2011. 水稻 HKT 基因家族密码子使用特性分析. 山西农业科学, 39 (11):1137-1140. | |
| [49] |
Yao G F, Ming M L, Allan A C, Gu C, Li L T, Wu X, Wang R Z, Chang Y J, Qi K J, Zhang S L, Wu J. 2017. Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant Journal, 92:437-451.
doi: 10.1111/tpj.2017.92.issue-3 URL |
| [50] |
Yu M, Man Y, Wang Y. 2019. Light-and temperature-induced expression of an R2R3-MYB gene regulates anthocyanin biosynthesis in red-fleshed kiwifruit. International Journal of Molecular Sciences, 20 (20):5228-5249.
doi: 10.3390/ijms20205228 URL |
| [51] |
Yue J Y, Liu J C, Tang W, Wu Y Q, Tang X F, Li W, Yang Y, Wang L H, Huang S X, Fang C B, Zhao K, Fei Z J, Liu Y S, Zheng Y. 2020. Kiwifruit Genome Database(KGD):a comprehensive resource for kiwifruit genomics. Horticulture Research, 7 (1):117-124.
doi: 10.1038/s41438-020-0338-9 |
| [52] | Zhang H, Li J, Wang R X, Zhi J K, Yin P, Xu J C. 2019. Comparative analysis of codon usage pattern of expansin genes from eight plant species. Journal of Biomolecular Structure & Dynamics, 37 (4):910-917. |
| [1] | WANG Quancheng, WU Jun, LI Lei, SHI Yanxia, XIE Xuewen, LI Baoju, CHAI Ali. Exploration on the Function of Pathogenicity-related Gene CcTLS1 in Corynespora cassiicola From Cucumber [J]. Acta Horticulturae Sinica, 2023, 50(3): 569-582. |
| [2] | WANG Xiaochen, NIE Ziye, LIU Xianju, DUAN Wei, FAN Peige, LIANG Zhenchang. Effects of Abscisic Acid on Monoterpene Synthesis in‘Jingxiangyu’Grape Berries [J]. Acta Horticulturae Sinica, 2023, 50(2): 237-249. |
| [3] | REN Fei, LU Miaomiao, LIU Jixiang, CHEN Xinli, LIU Daofeng, SUI Shunzhao, MA Jing. Expression and Adversity Resistance Analysis of a Late Embryogenesis Abundant Protein Gene CpLEA from Chimonanthus praecox [J]. Acta Horticulturae Sinica, 2023, 50(2): 359-370. |
| [4] | ZHAI Hanhan, ZHAI Yujie, TIAN Yi, ZHANG Ye, YANG Li, WEN Zhiliang, CHEN Haijiang. Genome-wide Identification of Peach SAUR Gene Family and Characterization of PpSAUR5 Gene [J]. Acta Horticulturae Sinica, 2023, 50(1): 1-14. |
| [5] | YUAN Xin, XU Yunhe, ZHANG Yupei, SHAN Nan, CHEN Chuying, WAN Chunpeng, KAI Wenbin, ZHAI Xiawan, CHEN Jinyin, GAN Zengyu. Studies on AcAREB1 Regulating the Expression of AcGH3.1 During Postharvest Ripening of Kiwifruit [J]. Acta Horticulturae Sinica, 2023, 50(1): 53-64. |
| [6] | LIN Haijiao, LIANG Yuchen, LI Ling, MA Jun, ZHANG Lu, LAN Zhenying, YUAN Zening. Exploration and Regulation Network Analysis of CBF Pathway Related Cold Tolerance Genes in Lavandula angustifolia [J]. Acta Horticulturae Sinica, 2023, 50(1): 131-144. |
| [7] | ZHAO Xueyan, WANG Qi, WANG Li, WANG Fangyuan, WANG Qing, LI Yan. Comparative Transcriptome Analysis of Differential Expression in Different Tissues of Corydalis yanhusuo [J]. Acta Horticulturae Sinica, 2023, 50(1): 177-187. |
| [8] | SONG Fang, CHEN Qi, YUAN Yanliang, CHEN Sha, YIN Haijun, and JIANG Yingchun, . A New Yellow-fleshed Kiwifruit Cultivar‘Xianwo 1’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 47-48. |
| [9] | QI Yongjie, GAO Zhenghui, MA Na, WANG Qingming, KE Fanjun, CHEN Qian, and XU Yiliu, . A New Yellow Flesh and Resistant to Canker Disease Kiwifruit Cultivar ‘Wannong Jinguo’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 49-50. |
| [10] | ZHANG Huiqin, LOU Guorong, LU Linghong, GU Xianbin, SONG Genhua, and XIE Ming. A New Yellow-fleshed Kiwifruit Cultivar‘Jinyi’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 51-52. |
| [11] | GAO Yanlong, WU Yuxia, ZHANG Zhongxing, WANG Shuangcheng, ZHANG Rui, ZHANG De, WANG Yanxiu. Bioinformatics Analysis of Apple ELO Gene Family and Its Expression Analysis Under Low Temperature Stress [J]. Acta Horticulturae Sinica, 2022, 49(8): 1621-1636. |
| [12] | QIU Ziwen, LIU Linmin, LIN Yongsheng, LIN Xiaojie, LI Yongyu, WU Shaohua, YANG Chao. Cloning and Functional Analysis of the MbEGS Gene from Melaleuca bracteata [J]. Acta Horticulturae Sinica, 2022, 49(8): 1747-1760. |
| [13] | TAO Xin, ZHU Rongxiang, GONG Xin, WU Lei, ZHANG Shaoling, ZHAO Jianrong, ZHANG Huping. Fructokinase Gene PpyFRK5 Plays an Important Role in Sucrose Accumulation of Pear Fruit [J]. Acta Horticulturae Sinica, 2022, 49(7): 1429-1440. |
| [14] | ZHENG Lin, WANG Shuai, LIU Yunuo, DU Meixia, PENG Aihong, HE Yongrui, CHEN Shanchun, ZOU Xiuping. Gene Cloning and Expression Analysis of NAC Gene in Citrus in Response to Huanglongbing [J]. Acta Horticulturae Sinica, 2022, 49(7): 1441-1457. |
| [15] | ZHANG Qiuyue, LIU Changlai, YU Xiaojing, YANG Jiading, FENG Chaonian. Screening of Reference Genes for Differentially Expressed Genes in Pyrus betulaefolia Plant Under Salt Stress by qRT-PCR [J]. Acta Horticulturae Sinica, 2022, 49(7): 1557-1570. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd