
Acta Horticulturae Sinica ›› 2023, Vol. 50 ›› Issue (3): 508-522.doi: 10.16420/j.issn.0513-353x.2021-1219
• Research Papers • Previous Articles Next Articles
YU Qinhan1, LI Junduo1, CUI Ying1, WANG Jiahui1, ZHENG Qiaoling1, XU Weirong1,2,3,4,*( )
)
Received:2022-08-18
Revised:2022-11-08
Online:2023-03-25
Published:2023-04-03
Contact:
*(E-mail:xuwr@nxu.edu.cn)
CLC Number:
YU Qinhan, LI Junduo, CUI Ying, WANG Jiahui, ZHENG Qiaoling, XU Weirong. Screening and Identification of Interacting Protein of VaMYB4a from Vitis amurensis[J]. Acta Horticulturae Sinica, 2023, 50(3): 508-522.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2021-1219
| 用途 Application | 引物编号 Primer No. | 引物序列(5′-3′) Primer sequence |
|---|---|---|
| 构建载体 | pDONR222-F | TCCCAGTCACGACGTTGTAAAACGACGGCCAGTCTT |
| Vector construction | pDONR222-R | AGAGCTGCCAGGAAACAGCTATGACCATGTAATACGACTC |
| BD-VaMYB4a-F | atggccatggaggccgaattcATGGGGAGGTCTCCTTGCTG | |
| BD-VaMYB4a-R | ccgctgcaggtcgacggatccTTATTTCATCTCCAAACCTCTGTAATC | |
| BD-VaMYB4a1-112-F | atggccatggaggccgaattcATGGGGAGGTCTCCTTGCTG | |
| BD-VaMYB4a1-112-R | ctagttatgcggccgctgcagCTTTCTCCGTATGTGGGTGTTCC | |
| BD-VaMYB4a113-252-F | atggccatggaggccgaattcAGCTTCTGAACCGAGGCATCG | |
| BD-VaMYB4a113-252-R | ctagttatgcggccgctgcagTTATTTCATCTCCAAACCTCTGTAATC | |
| AD-VaCOL2-F | gccatggaggccagtgaattcATGTTGAAGGACGAAGGTTGTAACG | |
| AD-VaCOL2-R | cagctcgagctcgatggatccTTAGAATGACGGGACAATGCC | |
| AD-VaCOL4-F | gccatggaggccagtgaattcATGGTTGTTGAAGTGGAAAGTTGG | |
| AD-VaCOL4-R | cagctcgagctcgatggatccTCAAAACGATGGAACGACGC | |
| AD-VaCOL5-F | gccatggaggccagtgaattcATGGGATTCCAGGAACTTAACG | |
| AD-VaCOL5-R | cagctcgagctcgatggatccTTAATAGGACGGAACGACACCG | |
| pBI221CE-VaMYB4a-F | gagaacacgggggactctagaATGGATGCAGCTAAGGACGG | |
| pBI221CE-VaMYB4a-R | aaattcggtaccccgctcgagTTATTTCATCTCCAAACCTCTGTAATC | |
| pBI221CE-VaMYB4a1-112-F | gagaacacgggggactctagaATGGATGCAGCTAAGGACGG | |
| pBI221CE-VaMYB4a1-112-R | aaattcggtaccccgctcgagTCTCCGTATGTGGGTGTTCCA | |
| pBI221CE-VaMYB4a113-252-F | gagaacacgggggactctagaAAGCTTCTGAACCGAGGCATC | |
| pBI221CE-MYB4a113-252-R | aaattcggtaccccgctcgagTTATTTCATCTCCAAACCTCTGTAATC | |
| pBI221NE-COL2-F | cccaggcctactagtggatccATGTTGAAGGACGAAGGTTGTAACG | |
| pBI221NE-COL2-R | cccgggagcggtaccctcgagGAATGACGGGACAATGCC | |
| pBI221NE-COL4-F | cccaggcctactagtggatccATGGTTGTTGAAGTGGAAAGTTGG | |
| pBI221NE-COL4-R | cccgggagcggtaccctcgagTCAAAACGATGGAACGACGC | |
| pBI221NE-VaCOL5-F | cccaggcctactagtggatccATGGGATTCCAGGAACTTAACG | |
| pBI221NE-VaCOL5-R | cccgggagcggtaccctcgagATAGGACGGAACGACACCG | |
| 实时荧光定量PCR | VaMYB4a-RT-F | TCGCTTATATCCGGGCACAC |
| Quantitative Real-time PCR | VaMYB4a-RT-R | AGGTCAGGCCTCAGGTAGTT |
| VaCOL2-RT-F | TTACAGCGTTCCGCACAAGA | |
| VaCOL2-RT-R | CCCCAGATGAAAACCCTGCT | |
| VaCOL4-RT-F | GGTGAACGACAACTGCTTTG | |
| VaCOL4-RT-R | GCACGACTCCCACTTCTAAA | |
| VvActin01-F | CTATCCTTCGTCTTGACCTTGCTG | |
| VvActin01-R | AGTGGTGAACATGTAACCCCTCTC | |
| PCR检测 | REC-F | GATGATGAAGATACCCCACCAAACCC |
| PCR detection | REC-R | GGTGCACGATGCACAGTTGAAGTG |
Table 1 Primers used in this study
| 用途 Application | 引物编号 Primer No. | 引物序列(5′-3′) Primer sequence |
|---|---|---|
| 构建载体 | pDONR222-F | TCCCAGTCACGACGTTGTAAAACGACGGCCAGTCTT |
| Vector construction | pDONR222-R | AGAGCTGCCAGGAAACAGCTATGACCATGTAATACGACTC |
| BD-VaMYB4a-F | atggccatggaggccgaattcATGGGGAGGTCTCCTTGCTG | |
| BD-VaMYB4a-R | ccgctgcaggtcgacggatccTTATTTCATCTCCAAACCTCTGTAATC | |
| BD-VaMYB4a1-112-F | atggccatggaggccgaattcATGGGGAGGTCTCCTTGCTG | |
| BD-VaMYB4a1-112-R | ctagttatgcggccgctgcagCTTTCTCCGTATGTGGGTGTTCC | |
| BD-VaMYB4a113-252-F | atggccatggaggccgaattcAGCTTCTGAACCGAGGCATCG | |
| BD-VaMYB4a113-252-R | ctagttatgcggccgctgcagTTATTTCATCTCCAAACCTCTGTAATC | |
| AD-VaCOL2-F | gccatggaggccagtgaattcATGTTGAAGGACGAAGGTTGTAACG | |
| AD-VaCOL2-R | cagctcgagctcgatggatccTTAGAATGACGGGACAATGCC | |
| AD-VaCOL4-F | gccatggaggccagtgaattcATGGTTGTTGAAGTGGAAAGTTGG | |
| AD-VaCOL4-R | cagctcgagctcgatggatccTCAAAACGATGGAACGACGC | |
| AD-VaCOL5-F | gccatggaggccagtgaattcATGGGATTCCAGGAACTTAACG | |
| AD-VaCOL5-R | cagctcgagctcgatggatccTTAATAGGACGGAACGACACCG | |
| pBI221CE-VaMYB4a-F | gagaacacgggggactctagaATGGATGCAGCTAAGGACGG | |
| pBI221CE-VaMYB4a-R | aaattcggtaccccgctcgagTTATTTCATCTCCAAACCTCTGTAATC | |
| pBI221CE-VaMYB4a1-112-F | gagaacacgggggactctagaATGGATGCAGCTAAGGACGG | |
| pBI221CE-VaMYB4a1-112-R | aaattcggtaccccgctcgagTCTCCGTATGTGGGTGTTCCA | |
| pBI221CE-VaMYB4a113-252-F | gagaacacgggggactctagaAAGCTTCTGAACCGAGGCATC | |
| pBI221CE-MYB4a113-252-R | aaattcggtaccccgctcgagTTATTTCATCTCCAAACCTCTGTAATC | |
| pBI221NE-COL2-F | cccaggcctactagtggatccATGTTGAAGGACGAAGGTTGTAACG | |
| pBI221NE-COL2-R | cccgggagcggtaccctcgagGAATGACGGGACAATGCC | |
| pBI221NE-COL4-F | cccaggcctactagtggatccATGGTTGTTGAAGTGGAAAGTTGG | |
| pBI221NE-COL4-R | cccgggagcggtaccctcgagTCAAAACGATGGAACGACGC | |
| pBI221NE-VaCOL5-F | cccaggcctactagtggatccATGGGATTCCAGGAACTTAACG | |
| pBI221NE-VaCOL5-R | cccgggagcggtaccctcgagATAGGACGGAACGACACCG | |
| 实时荧光定量PCR | VaMYB4a-RT-F | TCGCTTATATCCGGGCACAC |
| Quantitative Real-time PCR | VaMYB4a-RT-R | AGGTCAGGCCTCAGGTAGTT |
| VaCOL2-RT-F | TTACAGCGTTCCGCACAAGA | |
| VaCOL2-RT-R | CCCCAGATGAAAACCCTGCT | |
| VaCOL4-RT-F | GGTGAACGACAACTGCTTTG | |
| VaCOL4-RT-R | GCACGACTCCCACTTCTAAA | |
| VvActin01-F | CTATCCTTCGTCTTGACCTTGCTG | |
| VvActin01-R | AGTGGTGAACATGTAACCCCTCTC | |
| PCR检测 | REC-F | GATGATGAAGATACCCCACCAAACCC |
| PCR detection | REC-R | GGTGCACGATGCACAGTTGAAGTG |
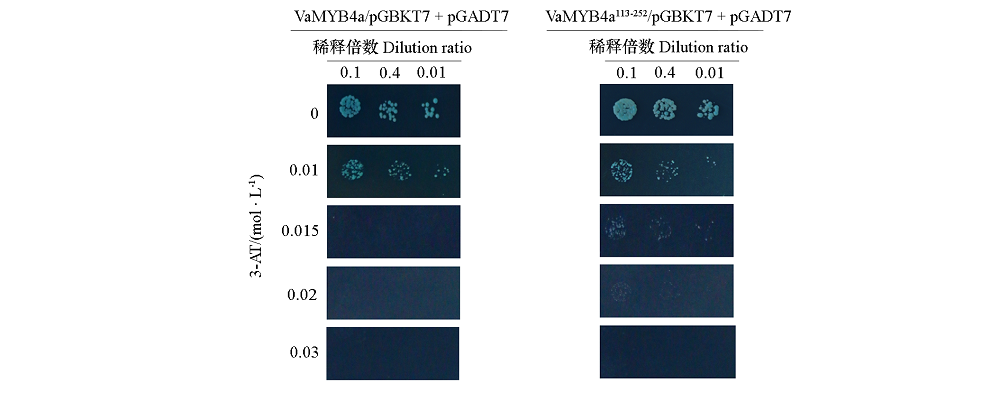
Fig. 2 Inhibition self-activation of VaMYB4a/pGBKT7 and VaMYB4a113-252/pGBKT7 on SD/-Trp/-Leu/-His/-Ade/AbA/X-α-gal medium with different concentrations of 3-AT

Fig. 3 Yeast two-hybrid screening of VaMYB4a113-252 interacting-proteins A:The growth situation of yeast;B:PCR identification of candidate clones(M:DL5000 marker,1-33:Positive clones;34:Negative control).
| 序号 Clone No. | NCBI比对结果 NCBI comparison results | E值 E-value | 相似度/% Identity | 登录号 Accession |
|---|---|---|---|---|
| VaMYB4a113-252-1 | dTDP-4-脱氢甘露糖还原酶 dTDP-4-dehydrorhamnose reductase | 6.00E-103 | 64 | XP_002282339.1 |
| VaMYB4a113-252-2 | Nudix水解酶2 Nudix hydrolase 2 | 1.00E-160 | 100 | RVW33646.1 |
| VaMYB4a113-252-3 | 抗锈蛋白激酶Lr10 Rust resistance kinase Lr10 | 9.00E-179 | 77 | RVW52296.1 |
| VaMYB4a113-252-4 | 无活性的亮氨酸富集重复类受体蛋白激酶 PREDICTED:probably inactive leucine-rich repeat receptor-like protein kinase | 0.0 | 82 | XP_003635622.1 |
| VaMYB4a113-252-5 | BEL1-like同源蛋白2 BEL1-like homeodomain protein 2 | 5e-61 | 100 | XP_034683092.1 |
| VaMYB4a113-252-6 | 类几丁质酶蛋白2 Chitinase-like protein 2 | 1.00E-161 | 100 | RVW69527.1 |
| VaMYB4a113-252-7 | 紫色酸性磷酸酶 PREDICTED:purple acid phosphatase 3 | 0.0 | 99 | XP_002285160.1 |
| VaMYB4a113-252-8 | 锌指蛋白 CONSTANS-like 4 Zinc finger protein CONSTANS-like 4 | 9.00E-141 | 100 | RVW58188.1 |
| VaMYB4a113-252-9 | 噻胺噻唑合成酶1,叶绿体 Thiamine thiazole synthase 1,chloroplastic | 0.0 | 91 | XP_002281769.1 |
| VaMYB4a113-252-10 | 半乳糖醛酸转移酶13亚型X1 Probable galacturonosyltransferase 13 isoform X1 | 0.0 | 99 | XP_034708194.1 |
| VaMYB4a113-252-11 | 蛋白质S-酰基转移酶24 Protein S-acyltransferase 24 | 0.0 | 92 | XP_002262878.1 |
| VaMYB4a113-252-12 | ABC转运蛋白F家族成员1 ABC transporter F family member 1 | 6.00E-166 | 86 | XP_010651804.1 |
| VaMYB4a113-252-13 | 萌发特异性半胱氨酸蛋白酶 Germination-specific cysteine protease 1 | 0.0 | 87 | XP_010651804.1 |
| VaMYB4a113-252-14 | 过氧化氢酶同工酶1 PREDICTED:catalase isozyme 1 | 4.00E-168 | 86 | XP_010663966.1 |
| VaMYB4a113-252-15 | mRNA前体切割因子Im 25kDa亚基2亚型X1 PREDICTED:pre-mRNA cleavage factor Im 25 kDa subunit 2 isoform X1 | 2.00E-167 | 99 | XP_002267257.1 |
| VaMYB4a113-252-16 | GDP-L-半乳糖磷酸化酶1 GDP-L-galactose phosphorylase 1 | 3.00E-31 | 57 | RVW30017.1 |
| VaMYB4a113-252-17 | 锌指蛋白CONSTANS-like 5 Zinc finger protein CONSTANS-like 5 | 1.00E-163 | 75 | RVW79681.1 |
Table 2 BLAST analysis of candidate proteins interacted with VaMYB4a113-252 in the Y2H system
| 序号 Clone No. | NCBI比对结果 NCBI comparison results | E值 E-value | 相似度/% Identity | 登录号 Accession |
|---|---|---|---|---|
| VaMYB4a113-252-1 | dTDP-4-脱氢甘露糖还原酶 dTDP-4-dehydrorhamnose reductase | 6.00E-103 | 64 | XP_002282339.1 |
| VaMYB4a113-252-2 | Nudix水解酶2 Nudix hydrolase 2 | 1.00E-160 | 100 | RVW33646.1 |
| VaMYB4a113-252-3 | 抗锈蛋白激酶Lr10 Rust resistance kinase Lr10 | 9.00E-179 | 77 | RVW52296.1 |
| VaMYB4a113-252-4 | 无活性的亮氨酸富集重复类受体蛋白激酶 PREDICTED:probably inactive leucine-rich repeat receptor-like protein kinase | 0.0 | 82 | XP_003635622.1 |
| VaMYB4a113-252-5 | BEL1-like同源蛋白2 BEL1-like homeodomain protein 2 | 5e-61 | 100 | XP_034683092.1 |
| VaMYB4a113-252-6 | 类几丁质酶蛋白2 Chitinase-like protein 2 | 1.00E-161 | 100 | RVW69527.1 |
| VaMYB4a113-252-7 | 紫色酸性磷酸酶 PREDICTED:purple acid phosphatase 3 | 0.0 | 99 | XP_002285160.1 |
| VaMYB4a113-252-8 | 锌指蛋白 CONSTANS-like 4 Zinc finger protein CONSTANS-like 4 | 9.00E-141 | 100 | RVW58188.1 |
| VaMYB4a113-252-9 | 噻胺噻唑合成酶1,叶绿体 Thiamine thiazole synthase 1,chloroplastic | 0.0 | 91 | XP_002281769.1 |
| VaMYB4a113-252-10 | 半乳糖醛酸转移酶13亚型X1 Probable galacturonosyltransferase 13 isoform X1 | 0.0 | 99 | XP_034708194.1 |
| VaMYB4a113-252-11 | 蛋白质S-酰基转移酶24 Protein S-acyltransferase 24 | 0.0 | 92 | XP_002262878.1 |
| VaMYB4a113-252-12 | ABC转运蛋白F家族成员1 ABC transporter F family member 1 | 6.00E-166 | 86 | XP_010651804.1 |
| VaMYB4a113-252-13 | 萌发特异性半胱氨酸蛋白酶 Germination-specific cysteine protease 1 | 0.0 | 87 | XP_010651804.1 |
| VaMYB4a113-252-14 | 过氧化氢酶同工酶1 PREDICTED:catalase isozyme 1 | 4.00E-168 | 86 | XP_010663966.1 |
| VaMYB4a113-252-15 | mRNA前体切割因子Im 25kDa亚基2亚型X1 PREDICTED:pre-mRNA cleavage factor Im 25 kDa subunit 2 isoform X1 | 2.00E-167 | 99 | XP_002267257.1 |
| VaMYB4a113-252-16 | GDP-L-半乳糖磷酸化酶1 GDP-L-galactose phosphorylase 1 | 3.00E-31 | 57 | RVW30017.1 |
| VaMYB4a113-252-17 | 锌指蛋白CONSTANS-like 5 Zinc finger protein CONSTANS-like 5 | 1.00E-163 | 75 | RVW79681.1 |

Fig. 4 Analysis of the interaction between VaMYB4a,VaCOL4 and VaCOL5 by Y2H assay(A)and BiFC assay(B) DDO:SD/-Trp/-Leu;QDO/X/A/3-AT:SD/-Trp/-Leu/-His/-Ade/X-α-gal/3-AT.
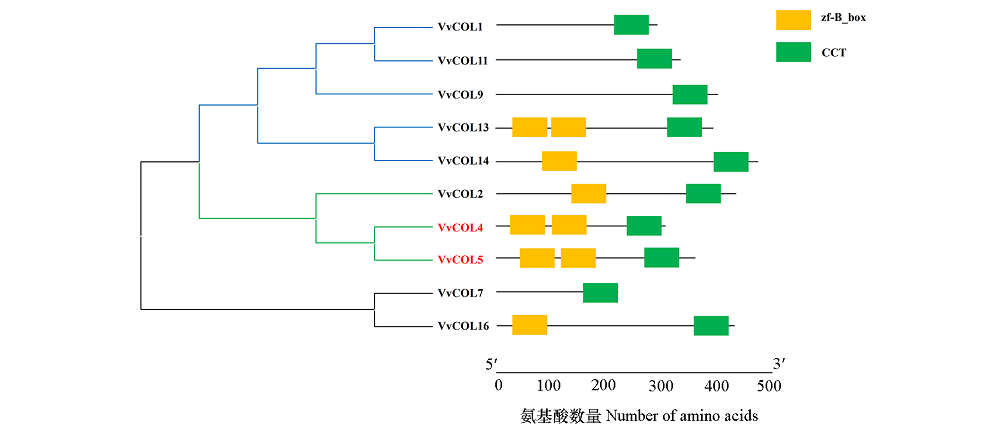
Fig. 5 The phylogenetic and conserved domains analysis of VvCOL proteins MEGA 5 with 1 000 bootstrap replicates. VvCOL1:XM_002268532.2;VvCOL11:XM_019223312.1;VvCOL9:XM_002264470.4;VvCOL13:KX986754.1;VvCOL14:XM_002263422.3;VvCOL2:XM_002282473.4;VvCOL4:XM_002263422.3;VvCOL7:XM_002274643.4;VvCOL16:XM_002282542.4.
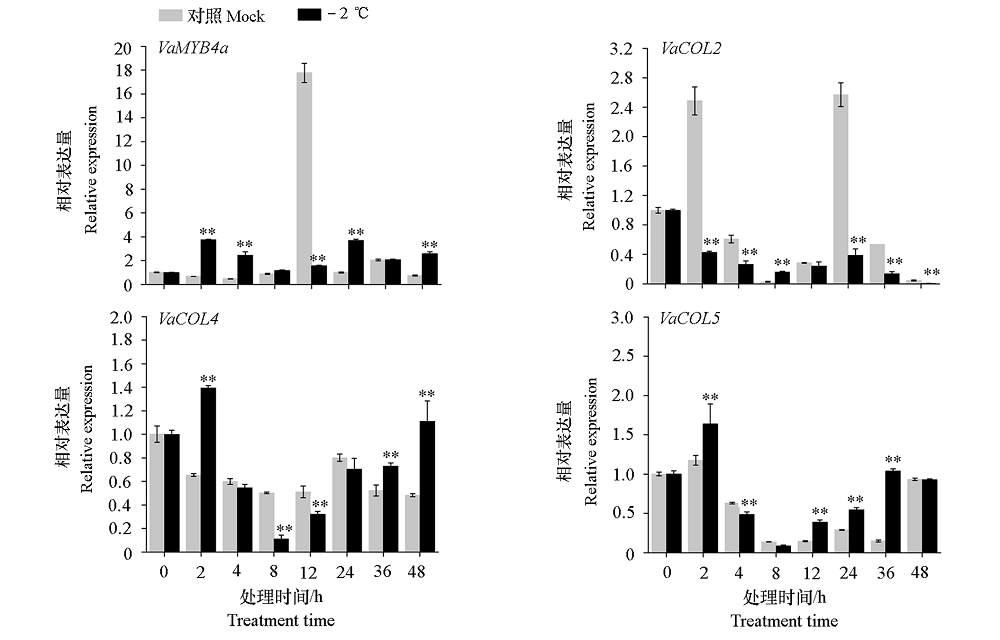
Fig. 8 Analysis of expression patterns of VaMYB4a and its interaction protein under low temperature **indicated significant difference compared with control groups(α = 0.01).
| [1] | Agarwal M, Hao Y J, Kapoor A, Dong C H, Fujii H, Zheng X, Zhu J K. 2006. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. Journal of Biological Chemistry, 281 (49):37635-37645. |
| [2] |
An J P, An X H, Yao J F, Wang X N, You C X, Wang X F, Hao Y J. 2018a. BTB protein MdBT 2 inhibits anthocyanin and proanthocyanidin biosynthesis by triggering MdMYB9 degradation in apple. Tree Physiology, 38 (10):1578-1587.
doi: 10.1093/treephys/tpy063 URL |
| [3] |
An J P, Li R, Qu F J, You C X, Wang X F, Hao Y J. 2018b. R2R3-MYB transcription factor MdMYB 23 is involved in the cold tolerance and proanthocyanidin accumulation in apple. The Plant Journal, 96 (3):562-577.
doi: 10.1111/tpj.14050 URL |
| [4] |
An J P, Wang X F, Espley R V, Wang K L, Bi S Q, You C X, Hao Y J. 2020a. An apple B-box protein MdBBX37 modulates anthocyanin biosynthesis and hypocotyl elongation synergistically with MdMYBs and MdHY5. Plant Cell Physiology, 61 (1):130-143.
doi: 10.1093/pcp/pcz185 URL |
| [5] |
An J P, Wang X F, Zhang X W, Xu H F, Bi S Q, You C X, Hao Y J. 2020b. An apple MYB transcription factor regulates cold tolerance and anthocyanin accumulation and undergoes MIEL1-mediated degradation. Plant Biotechnology Journal, 18 (2):337-353.
doi: 10.1111/pbi.v18.2 URL |
| [6] | An J P, Wang X F, Zhang X W, You C X, Hao Y J. 2021. Apple B-box protein BBX37 regulates jasmonic acid mediated cold tolerance through the JAZ-BBX37-ICE1-CBF pathway and undergoes MIEL1-mediated ubiquitination and degradation. New Phytologist, 229 (5):2709-2729. |
| [7] |
Bai S, Tao R, Tang Y, Yin L, Ma Y, Ni J, Yan X, Yang Q, Wu Z, Zeng Y, Teng Y. 2019. BBX16,a B-box protein,positively regulates light-induced anthocyanin accumulation by activating MYB 10 in red pear. Plant Biotechnology Journal, 17 (10):1985-1997.
doi: 10.1111/pbi.v17.10 URL |
| [8] |
Cáceres-Mella A, Talaverano M I, Villalobos-González L, Ribalta-Pizarro C, Pastenes C. 2017. Controlled water deficit during ripening affects proanthocyanidin synthesis,concentration and composition in Cabernet Sauvignon grape skins. Plant Physiology Biochemistry, 117:34-41.
doi: 10.1016/j.plaphy.2017.05.015 URL |
| [9] |
Chen P, Zhi F, Li X, Shen W, Yan M, He J, Chana B, Fan T, Zhou S, Ma F. 2022. Zinc-finger protein MdBBX7/MdCOL9,a target of MdMIEL 1 E3 ligase,confers drought tolerance in apple. Plant Physiology, 188 (1):540-559.
doi: 10.1093/plphys/kiab420 URL |
| [10] | Chen Rugang, Gong Zhenhui, Lu Minghui, Li Dawei, Huang Wei. 2010. Research advanced of the transcription factors networks related to plant adverse environmental stress. Journal of Agricultural Biotechnology, 18 (1):126-134. (in Chinese) |
| 陈儒钢, 巩振辉, 逯明辉, 李大伟, 黄炜. 2010. 植物抗逆反应中的转录因子网络研究进展. 农业生物技术学报, 18 (1):126-134. | |
| [11] | Chen Y, Chen Z, Kang J, Kang D M, Gu H, Qin G. 2013. AtMYB14 regulates cold tolerance in Arabidopsis. Plant molecular Biology Report, 31 (1):87-97. |
| [12] |
Cheng X F, Wang Z Y. 2005. Overexpression of COL9,a CONSTANS-LIKE gene,delays flowering by reducing expression of CO and FT in Arabidopsis thaliana. The Plant Journal, 43 (5):758-768.
doi: 10.1111/tpj.2005.43.issue-5 URL |
| [13] |
Cheo D L, Titus S A, Byrd D R, Hartley J L, Temple G F, Brasch M A. 2004. Concerted assembly and cloning of multiple DNA segments using in vitro site-specific recombination:functional analysis of multi-segment expression clones. Genome Research, 14 (10b):2111-2120.
doi: 10.1101/gr.2512204 URL |
| [14] |
Cho L H, Yoon J, An G. 2017. The control of flowering time by environmental factors. The Plant Journal, 90 (4):708-719.
doi: 10.1111/tpj.2017.90.issue-4 URL |
| [15] |
Czemmel S, Stracke R, Weisshaar B, Cordon N, Harris N N, Walker A R, Robinson S P, Bogs J. 2009. The grapevine R2R3-MYB transcription factor VvMYBF 1 regulates flavonol synthesis in developing grape berries. Plant Physiology, 151 (3):1513-1530.
doi: 10.1104/pp.109.142059 pmid: 19741049 |
| [16] |
Edery I, Chu L L, Sonenberg N, Pelletier J. 1995. An efficient strategy to isolate full-length cDNAs based on an mRNA cap retention procedure (CAPture). Molecular Cellular Biology, 15 (6):3363-3371.
doi: 10.1128/MCB.15.6.3363 URL |
| [17] |
Gangappa S N, Botto J F. 2014. The BBX family of plant transcription factors. Trends in Plant Science, 19 (7):460-470.
doi: 10.1016/j.tplants.2014.01.010 pmid: 24582145 |
| [18] | Hou S, Zhao T, Yang D, Li Q, Liang L, Wang G, Ma Q. 2021. Selection and validation of reference genes for quantitative RT-PCR analysis in Corylus heterophylla Fisch. × Corylus avellana L. Plants (Basel), 10 (1):159. |
| [19] |
Katzen F. 2007. Gateway recombinatorial cloning:a biological operating system. Expert Opinion on Drug Discovery, 2 (4):571-589.
doi: 10.1517/17460441.2.4.571 URL |
| [20] |
Landia M, Tattinib M, Gould K S. 2015. Multiple functional roles of anthocyanins in plant-environment interactions. Environmental Experimental Botany, 119:4 -17.
doi: 10.1016/j.envexpbot.2015.05.012 URL |
| [21] |
Lee H G, Seo P J. 2016. The Arabidopsis MIEL 1 E3 ligase negatively regulates ABA signalling by promoting protein turnover of MYB96. Nature Communications, 7:12525.
doi: 10.1038/ncomms12525 |
| [22] | Li C, Pei J, Yan X, Cui X, Tsuruta M, Liu Y, Lian C. 2021a. A poplar B-box protein PtrBBX23 modulates the accumulation of anthocyanins and proanthocyanidins in response to high light. Plant Cell & Environment, 44 (9):3015-3033. |
| [23] |
Li Y, Shi Y, Li M, Fu D, Wu S, Li J, Gong Z, Liu H, Yang S. 2021b. The CRY2-COP1-HY5-BBX7/ 8 module regulates blue light-dependent cold acclimation in Arabidopsis. The Plant Cell, 33 (11):3555-3573.
doi: 10.1093/plcell/koab215 URL |
| [24] |
Liu Y, Lin G, Yin C M, Fang Y. 2020. B-box transcription factor 28 regulates flowering by interacting with constans. Scientific Reports, 10 (1):17889.
doi: 10.1038/s41598-020-74838-8 |
| [25] |
Lu Suwen, Zheng Xuanang, Wang Jiayang, Fang Jinggui. 2021. Research progress on the metabolism of flavonoids in grape. Acta Horticulturae Sinica, 48 (12):2506-2524. (in Chinese)
doi: 10.16420/j.issn.0513-353x.2021-0558 URL |
|
卢素文, 郑暄昂, 王佳洋, 房经贵. 2021. 葡萄类黄酮代谢研究进展. 园艺学报, 48 (12):2506-2524.
doi: 10.16420/j.issn.0513-353x.2021-0558 URL |
|
| [26] |
Meng L, Fan Z, Qiang Z, Wang C, Gao Ying, Deng Y, Zhu B, Zhu H, Chen J, Shan W, Yin X, Zhong S, Grierson D, Jiang C Z, Luo Y, Fu D-Q. 2018. BEL1-LIKE HOMEODOMAIN 11 regulates chloroplast development and chlorophyll synthesis in tomato fruit. The Plant Journal, 94 (6):1126-1140.
doi: 10.1111/tpj.13924 pmid: 29659108 |
| [27] |
Merzlyak M N, Chivkunova O B. 2000. Light-stress-induced pigment changes and evidence for anthocyanin photoprotection in apples. Journal of Photochemistry Photobiology, 55 (2-3):155-163.
doi: 10.1016/S1011-1344(00)00042-7 URL |
| [28] |
Naing A H, Ai T N, Lim K B, Lee I J, Kim C K. 2018. Overexpression of Rosea 1 from snapdragon enhances anthocyanin accumulation and abiotic stress tolerance in transgenic tobacco. Frontiers in Plant Science, 9:1070.
doi: 10.3389/fpls.2018.01070 pmid: 30158941 |
| [29] |
Ogata K, Morikawa S, Nakamura H, Hojo H, Yoshimura S, Zhang R, Aimoto S, Ametani Y, Hirata Z, Sarai A. 1995. Comparison of the free and DNA-complexed forms of the DMA-binding domain from c-Myb. Nature Structural Biology, 2 (4):309-320.
doi: 10.1038/nsb0495-309 pmid: 7796266 |
| [30] | Pei Lili, Guo Yuhua, Xu Zhaoshi, Li Liancheng, Chen Ming, Ma Youzhi. 2012. Research progress on stress-related protein kinases in plants. Acta Botanica Boreali-Occidentalia Sinica, 32 (5):1052-1061. (in Chinese) |
| 裴丽丽, 郭玉华, 徐兆师, 李连城, 陈明, 马有志. 2012. 植物逆境胁迫相关蛋白激酶的研究进展. 西北植物学报, 32 (5):1052-1061. | |
| [31] | Peng Xiaoyu, Liu Rong, Yang Jing, Ding Xia, Wang Fang, Li Yangsheng, Cai Yaohui. 2014. Construction and identification of a mixed cDNA library of rice roots construction and identification of a mixed cDNA library of rice roots under salt and drought stress using Gateway technology. Chinese Journal of Applied and Environmental Biology,(2):291-294. (in Chinese) |
| 彭晓珏, 刘瑢, 阳菁, 丁霞, 王芳, 李阳生, 蔡耀辉. 2014. Gateway技术构建盐和干旱胁迫下水稻根系混合cDNA文库及其质量鉴定. 应用与环境生物学报,(2):291-294. | |
| [32] |
Piero A R L, Puglisi I, Rapisarda P, Petrone G. 2005. Anthocyanins accumulation and related gene expression in red orange fruit induced by low temperature storage. Journal of Agricultural Food Chemistry, 53 (23):9083-9088.
doi: 10.1021/jf051609s URL |
| [33] | Steinbach Y. 2019. The Arabidopsis thaliana CONSTANS-LIKE 4(COL4)- A modulator of flowering time. Frontiers in Plant Science, 28 (10):651. |
| [34] |
Su L T, Li J W, Liu D Q, Zhai Y, Zhang H J, Li X W, Zhang Q L, Wang Y, Wang Q Y. 2014. A novel MYB transcription factor,GmMYBJ1,from soybean confers drought and cold tolerance in Arabidopsis thaliana. Gene, 538 (1):46-55.
doi: 10.1016/j.gene.2014.01.024 URL |
| [35] |
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5:Molecular evolutionary genetics analysis using maximum likelihood,evolutionary distance,and maximum parsimony methods. Molecular Biology Evolution, 28 (10):2731-2739.
doi: 10.1093/molbev/msr121 URL |
| [36] |
Thompson M A, Ramsay R G. 1995. Myb:an old oncoprotein with new roles. Bioessays, 17 (4):341-350.
pmid: 7741726 |
| [37] |
Wang M, Hao J, Chen X, Zhang X. 2020. SlMYB 102 expression enhances low-temperature stress resistance in tomato plants. Peer J, 8:e10059.
doi: 10.7717/peerj.10059 URL |
| [38] | Wang Shun-li, Xue Jing-qi, Zhu Fu-yong, Zhang Ping, Ren Xiu-xia, Liu Chuan-jiao,and Zhang Xiu-xin. 2014. Molecular cloning,expression and evolutionary analysis of the floweringregulating transcription factor gene PsCOL4 in tree peony. Acta Horticulturae Sinica, 41 (7):1409-1417. (in Chinese) |
| 王顺利, 薛璟祺, 朱富勇, 张萍, 任秀霞, 刘传娇, 张秀新. 2014. 牡丹开花调控转录因子基因PsCOL4的克隆、表达与系统进化分析. 园艺学报, 41 (7):1409-1417. | |
| [39] |
Wang X F, An J P, Liu X, Su L, You C X, Hao Y J. 2018. The nitrate-responsive protein MdBT 2 regulates anthocyanin biosynthesis by interacting with the MdMYB1 transcription factor. Plant Physiology, 178 (2):890-906.
doi: 10.1104/pp.18.00244 URL |
| [40] |
Wang Y, Mao Z, Jiang H, Zhang Z, Chen X. 2019. A feedback loop involving MdMYB108L and MdHY 5 controls apple cold tolerance. Biochemical Biophysical Research Communications, 512 (2):381-386.
doi: 10.1016/j.bbrc.2019.03.101 URL |
| [41] |
Xie Y P, Chen P, Yan Y, Bao C, Li X W, Wang L, Shen X, Li H, Liu X, Niu C, Zhu C, Fang N, Shao Y, Zhao T, Yu J T, Zhu J H, Xu L F, Nocker S V, Ma F, Guan Q M. 2018. An atypical R2R3 MYB transcription factor increases cold hardiness by CBF-dependent and CBF-independent pathways in apple. New Phytologist, 218 (1):201-218.
doi: 10.1111/nph.14952 URL |
| [42] | Xing C, Liu Y, Zhao L, Zhang S, Huang X. 2019. A novel MYB transcription factor regulates ascorbic acid synthesis and affects cold tolerance. Plant,Cell & Environment, 42 (3):832-845. |
| [43] |
Yang A, Dai X, Zhang W H. 2012. A R2R3-type MYB gene,OsMYB2,is involved in salt,cold,and dehydration tolerance in rice. Journal of Experimental Botany, 63 (7):2541-2556.
doi: 10.1093/jxb/err431 pmid: 22301384 |
| [44] |
Yang T W, He Y, Niu S B, Yan S W, Zhang Y. 2020. Identification and characterization of the CONSTANS(CO)/CONSTANS-like(COL)genes related to photoperiodic signaling and flowering in tomato. Plant Science, 301:110653.
doi: 10.1016/j.plantsci.2020.110653 URL |
| [45] |
Yao C, Li X, Li Y, Yang G, Liu W, Shao B, Zhong J, Huang P, Han D. 2022. Overexpression of a Malus baccata MYB transcription factor gene MbMYB4 increases cold and drought tolerance in Arabidopsis thaliana. International Journal of Molecular Sciences, 23 (3):1794.
doi: 10.3390/ijms23031794 URL |
| [46] |
Yao W, Wang L, Wang J, Ma F, Wang Y. 2017. VpPUB24,a novel gene from Chinese grapevine,Vitis pseudoreticulata,targets VpICE 1 to enhance cold tolerance. Journal of Experimental Botany, 68 (11):2933-2949.
doi: 10.1093/jxb/erx136 URL |
| [47] | Yu Qin-han. 2022. Response mechanism of Amur grape VaMYB4a in response to cold stress[M. D. Dissertation]. Yinchuan: Ningxia University. (in Chinese) |
| 俞沁含. 2022. 山葡萄VaMYB4a参与低温胁迫应答的机理研究[硕士论文]. 银川: 宁夏大学. | |
| [48] |
Zhang H, Wang Z, Li X, Gao X, Dai Z, Cui Y, Zhi Y, Liu Q C, Zhai H, Gao S, Zhao N, He S Z. 2021. The IbBBX24-IbTOE3-IbPRX17 module enhances abiotic stress tolerance by scavenging reactive oxygen species in sweet potato. New Phytologist, 233 (3):1133-1152.
doi: 10.1111/nph.17860 pmid: 34773641 |
| [49] |
Zhang L C, Zhao G, Xia C, Jia J Z, Liu X, Kong X Y. 2012. Overexpression of a wheat MYB transcription factor gene,TaMYB56-B,enhances tolerances to freezing and salt stresses in transgenic Arabidopsis. Gene, 505 (1):100-107.
doi: 10.1016/j.gene.2012.05.033 URL |
| [50] |
Zhang Y T, Liu Y, Hu W, Sun B, Chen Q, Tang H. 2018. Anthocyanin accumulation and related gene expression affected by low temperature during strawberry coloration. Acta Physiologiae Plantarum, 40 (11):1-8.
doi: 10.1007/s11738-017-2577-4 |
| [51] | Zhu J H, Verslues P E, Zheng X W, Lee B-h, Zhan X Q, Manabe Y, Sokolchik I, Zhu Y, Dong C H, Zhu J K, Hasegawa P M, Bressan R A. 2005. HOS10 encodes an R2R3-type MYB transcription factor essential for cold acclimation in plants. Proceedings of the National Academy of Sciences, 102 (28):9966-9971. |
| [1] | MA Junjie, SONG Lina, LI Le, MA Xiaochun, JIN Lei, XU Weirong. VaCBL6 from Vitis amurensis Involved in Abiotic Stress Response and ABA-mediated Pathway [J]. Acta Horticulturae Sinica, 2021, 48(6): 1079-1093. |
| [2] | YU Qinhan, JIAO Shuzhen, WU Nan, ZHANG Ningbo, XU Weirong. Molecular Cloning,Expression and Polyclonal Antibody Preparation of E3 Ubiquitin Ligase Gene HOS1 from Vitis vinifera [J]. Acta Horticulturae Sinica, 2021, 48(6): 1173-1180. |
| [3] | ZHENG Qiaoling, SHEN Wei, YAO Wenkong, and XU Weirong, . Construction of Yeast Two-hybrid Three-frame cDNA Library of Vitis amurensis and Screening of VaCIPK18 Interaction Protein [J]. Acta Horticulturae Sinica, 2020, 47(12): 2301-2316. |
| [4] | ZHANG Huilin,ZHU Wan,TIAN Li,and ZHANG Wei*. Characterization and Expression Analysis of Petunia PhZPT2-12 Transcription Factor Related to Cold Response [J]. ACTA HORTICULTURAE SINICA, 2019, 46(8): 1543-1552. |
| [5] | ZHAO Yanqing1,DU Jianchang2,WANG Panqiao1,QIN Xiaodong1,and CHEN Jinfeng1,*. Identification and Expression Analysis of NAC Transcription Factor Gene Family Under Low Temperature in Cucumis sativus var. hardwickii [J]. ACTA HORTICULTURAE SINICA, 2019, 46(7): 1303-1319. |
| [6] | WANG Xiaodi,JI Xiaohao,ZHENG Xiaocui,WANG Yingying,SONG Yang*,and LIU Fengzhi*. Cloning and Functional Analysis of a Cold Treatment Response Factor Gene PdCIbHLH in Peach [J]. ACTA HORTICULTURAE SINICA, 2019, 46(3): 444-452. |
| [7] | XU Hongxia1,LI Xiaoying1,XU Changjie2,and CHEN Junwei1,*. A Study on Cold Resistance Function of Dehydrin Gene EjDHN5 from Loquat [J]. ACTA HORTICULTURAE SINICA, 2017, 44(2): 268-278. |
| [8] | WANG Hai-bo,CHENG Lai-liang,CHANG Yuan-sheng,SUN Qing-rong,TAO Ji-han,and LI Lin-guang. Physiological and Transcriptome Response of Apple Dwarfing Rootstock to Cold Stress and Cold-resistant Genes Screening [J]. ACTA HORTICULTURAE SINICA, 2016, 43(8): 1437-1451. |
| [9] | ZHOU Lin,CHEN Xuan,WANG Yu-hua,ZHU Xu-jun,LI Xing-hui,and FANG Wan-ping*. Effect of Harpin on Physiological Characteristics of Tea Plant Under Cold Stress [J]. ACTA HORTICULTURAE SINICA, 2014, 41(4): 746-754. |
| [10] | YANG Bing-Yan, HUO Xiu-Ai, LIU Yun-Ting, DUAN Hui-Jun. MSAP and Differential Expression of Homologous Diploid and Triploid Watermelon Under Cold Stress [J]. ACTA HORTICULTURAE SINICA, 2014, 41(11): 2313-2322. |
| [11] | BAI Su-Hua, ZHU Jun, DAI Hong-Yi. Cloning and Expression Analysis of a New Acyl-CoA-binding Protein 2 (MdACBP2)Identified from Malus domestica [J]. ACTA HORTICULTURAE SINICA, 2012, 39(10): 1893-1902. |
| [12] | XU Hong-xia;CHEN Jun-wei;YANG Yong;SUN Jun-wei;and YAN Cheng-qi . Isolation and Expression Analysis of DHN Gene in Loquat Fruit Under Cold Stress [J]. ACTA HORTICULTURAE SINICA, 2011, 38(6): 1071-1080. |
| [13] | HAO Qiang;ZHANG Rui-li;LENG Ping-sheng;and GUAN Xue-lian . Proteomic Analysis of Cold Stress Responses in Euonymus japonicus Leaves [J]. ACTA HORTICULTURAE SINICA, 2011, 38(11): 2169-2179. |
| [14] | CAO Yi-bin;SHI Rui;CHEN Wen-rong;and GUO Wei-dong . Comparative Analysis of Expression Changes of Ethylene Response Factor 6(ERF6)in Fingered Citron and Trifoliate Orange Under Cold Stress [J]. ACTA HORTICULTURAE SINICA, 2011, 38(10): 1873-1882. |
| [15] | LI Guang-qing;XIE Zhu-jie;YAO Xue-qin;and CHEN Xue-hao. Studies on the Relationship Between Chlorophyll Fluorescence Parameters and Cold Tolerance of Cauliflower [J]. ACTA HORTICULTURAE SINICA, 2010, 37(12): 2001-2001–2006. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd