
Acta Horticulturae Sinica ›› 2023, Vol. 50 ›› Issue (2): 359-370.doi: 10.16420/j.issn.0513-353x.2021-0987
• Research Papers • Previous Articles Next Articles
REN Fei, LU Miaomiao, LIU Jixiang, CHEN Xinli, LIU Daofeng, SUI Shunzhao, MA Jing*( )
)
Received:2022-09-03
Revised:2022-10-27
Online:2023-02-25
Published:2023-03-06
Contact:
*(E-mail:majing427@swu.edu.cn)
CLC Number:
REN Fei, LU Miaomiao, LIU Jixiang, CHEN Xinli, LIU Daofeng, SUI Shunzhao, MA Jing. Expression and Adversity Resistance Analysis of a Late Embryogenesis Abundant Protein Gene CpLEA from Chimonanthus praecox[J]. Acta Horticulturae Sinica, 2023, 50(2): 359-370.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2021-0987
| 基因 Gene | 正向引物(5′-3′) Forward primer | 反向引物(5′-3′) Reverse primer |
|---|---|---|
| CpLEA | CAGGAGAAGACGAGTCAGATGGT | CCGAGTCCCTTGTAGATTGG |
| Atactin | CTTCGTCTTCCACTTCAG | ATCATACCAGTCTCAACAC |
| Actin | GTTATGGTTGGGATGGGACAGAAAG | GGGCTTCAGTAAGGAAACAGGA |
| Tublin | TAGTGACAAGACAGTAGGTGGAGGT | GTAGGTTCCAGTCCTCACTTCATC |
Table 1 Primer sequences used in real-time quantitative PCR
| 基因 Gene | 正向引物(5′-3′) Forward primer | 反向引物(5′-3′) Reverse primer |
|---|---|---|
| CpLEA | CAGGAGAAGACGAGTCAGATGGT | CCGAGTCCCTTGTAGATTGG |
| Atactin | CTTCGTCTTCCACTTCAG | ATCATACCAGTCTCAACAC |
| Actin | GTTATGGTTGGGATGGGACAGAAAG | GGGCTTCAGTAAGGAAACAGGA |
| Tublin | TAGTGACAAGACAGTAGGTGGAGGT | GTAGGTTCCAGTCCTCACTTCATC |

Fig. 1 SDS-PAGE electrophoresis of CpLEA protein obtained by induction of Escherichia coli BL21/pET32a(+)-CpLEA M:Protein marker;1:Total protein without induction;2:Total protein after induction;3:Supernatant protein after induction;4:Precipitation protein after induction;5:Buffering solution;6,7,8:20,50,300 mmol · L-1 imidazole elution.

Fig. 2 Colony density(A)and bacterial growth curve(B)of Escherichia coli to CpLEA strain BL21/pET32a(+)-CpLEA and to empty vector strain BL21/pET32a(+)
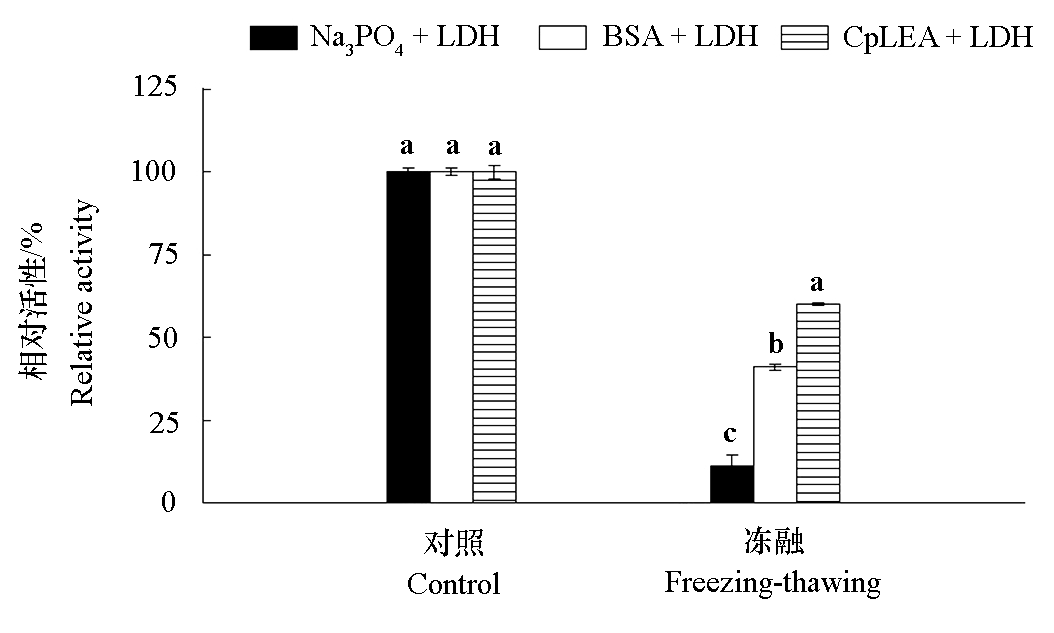
Fig. 3 Lactate dehydrogenase(LDH)activity by CpLEA protein under different stress conditions Different lowercase letters indicate significant differences among different reaction systems at 0.05 level.
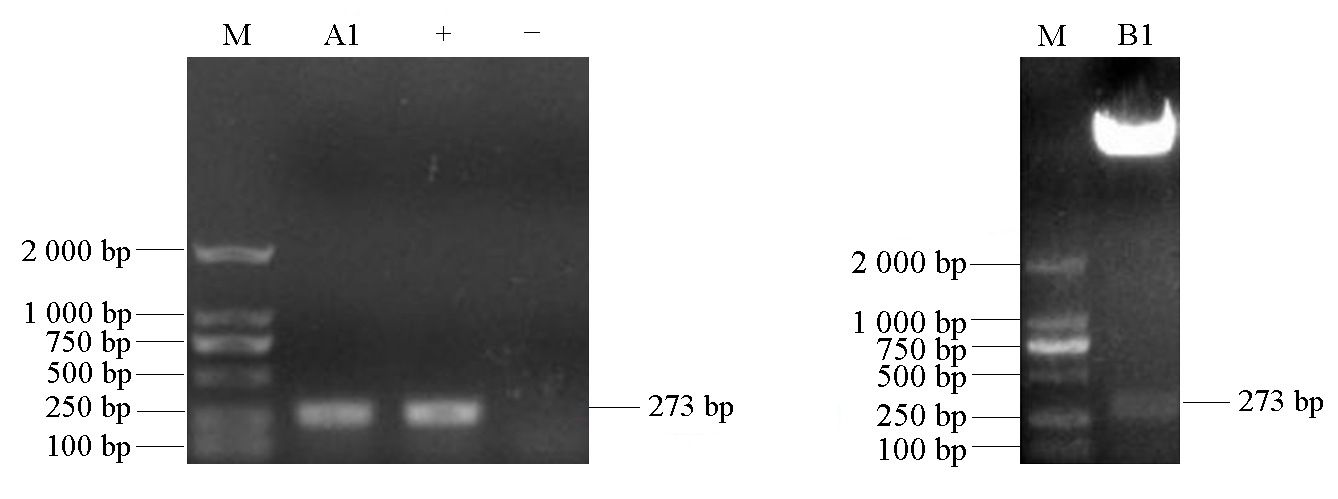
Fig. 4 PCR(A1)and enzyme cutting verification(B1)electrophoresis of plant expression vector M:DL2000 marker;+:Positive controlar;-:Negative control,ddH2O.

Fig. 5 PCR(A)and RT-PCR(B)of Arabidopsis thaliana overexpressing CpLEA lines and expression analysis of CpLEA(C) M:DL2000 marker;1-11,L-1-L-9:Transgenic lines;+:Positive controlar;-:Negative control,ddH2O;WT:Wild type.

Fig. 6 Drought resistance in Arabidopsis thaliana overexpressing CpLEA lines(L-6,L-7,L-4)and wild type(WT) A:Seed germination under 10% PEG simulated drought;B:Seedling phenotype after drought stress;C:Survival rate after rewatering;D:CMS after drought stress. Different lowercase letters indicate significant differences among materials at 0.05 level.
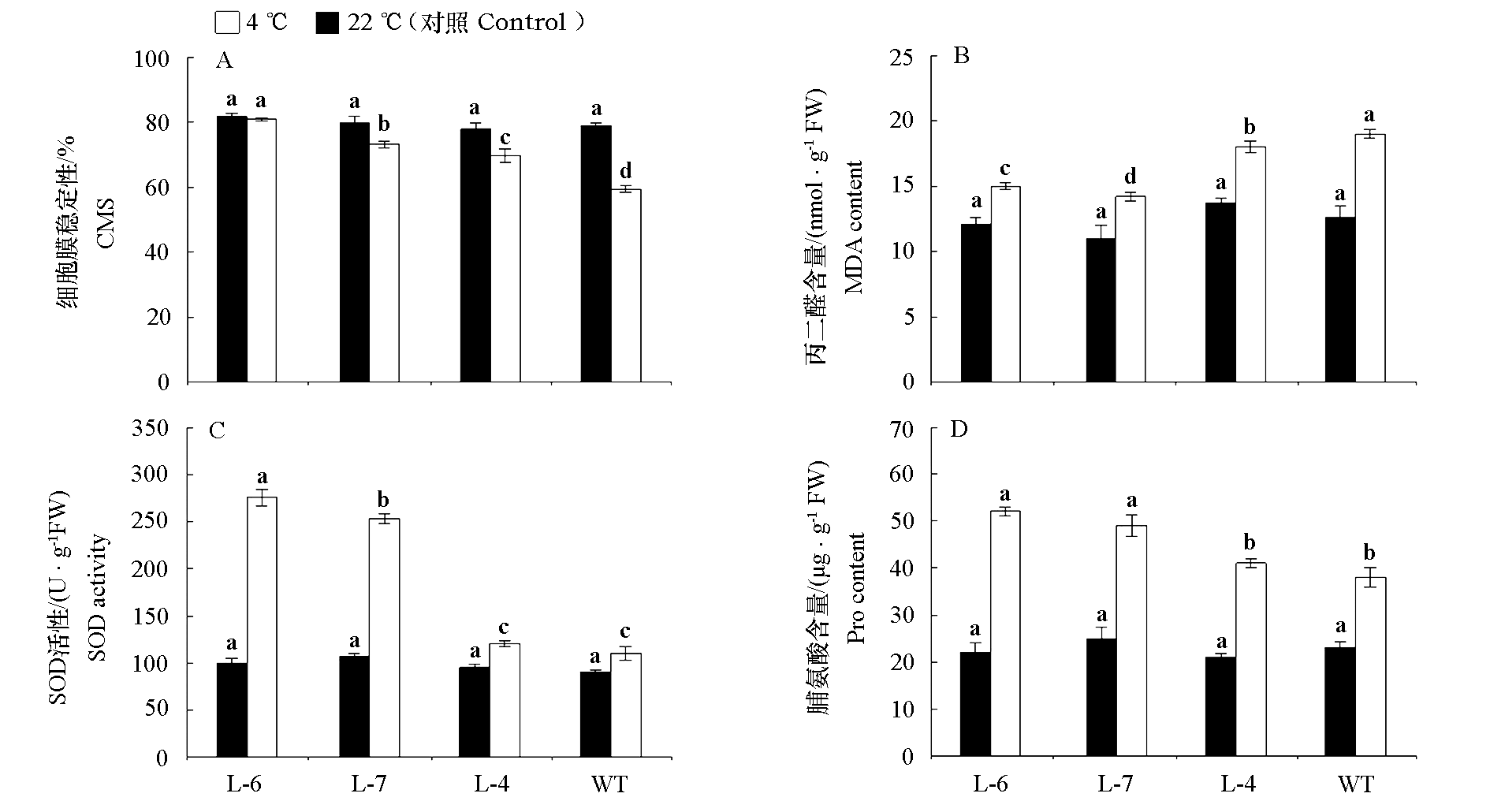
Fig. 8 Cold resistance in Arabidopsis thaliana overexpressing CpLEA lines(L-6,L-7,L-4)and wild type(WT) Different lowercase letters indicate significant differences among different materials with the same treatment at 0.05 level.
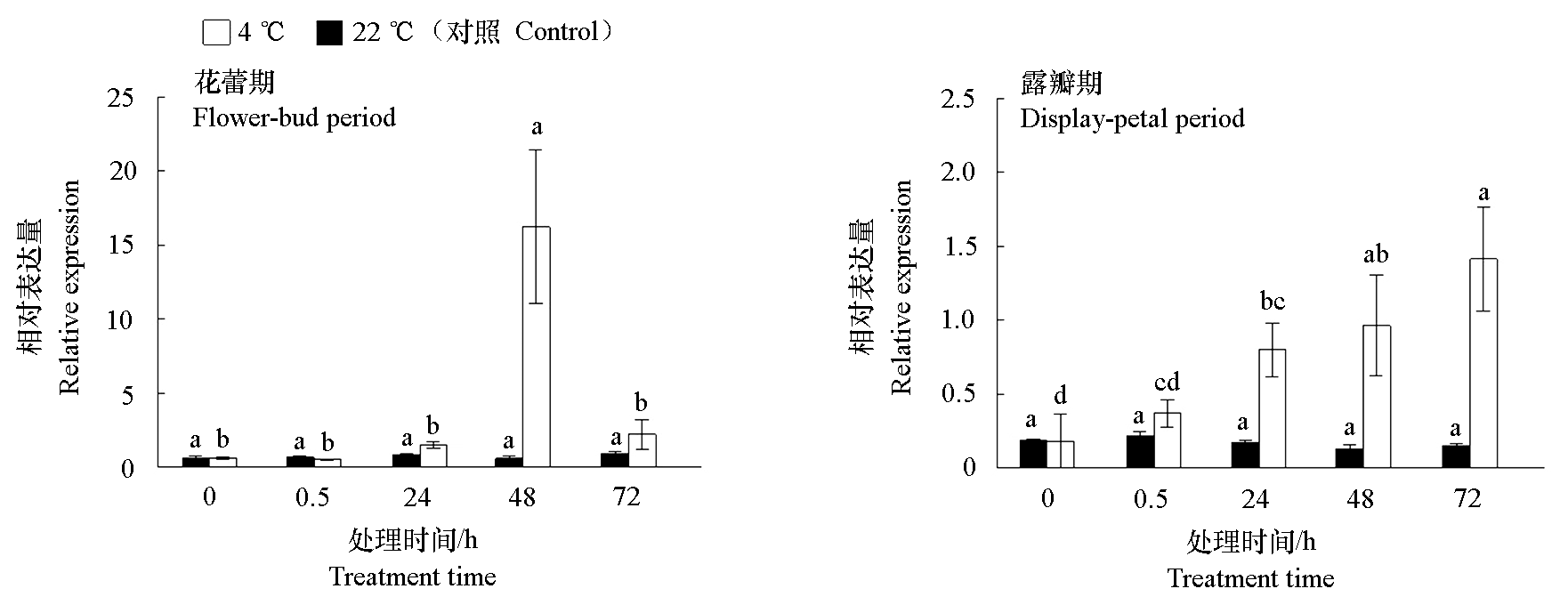
Fig. 9 Expression analysis of CpLEA during flower stage Different lowercase letters indicate significant differences in the same treatment and different treatment times at 0.05 level.
| [1] |
Babu R C, Zhang J X, Blum A, Ho D T, Wu R, Nguyen H. 2004. HVA1,a LEA gene from barley confers dehydration tolerance in transgenic rice(Oryza sativa L.)via cell membrane protection. Plant Science, 166 (4):855-862.
doi: 10.1016/j.plantsci.2003.11.023 URL |
| [2] | Bao F, Du D L, An Y, Yang W R, Wang J, Cheng T R, Zhang Q X. 2017. Overexpression of Prunus mume dehydrin genes in tobacco enhances tolerance to cold and drought. Frontiers in Plant Science, 8:151-163. |
| [3] |
Battaglia M, Olvera-Carrillo Y, Garciarrubio A, Campos F, Covarrubias A A. 2008. The enigmatic LEA proteins and other hydrophilins. Plant Physiol, 148 (1):6-24.
doi: 10.1104/pp.108.120725 pmid: 18772351 |
| [4] |
Chen J, Fan L, Du Y, Zhu W N, Tang Z Q, Li N, Zhang D P, Zhang L S. 2016. Temporal and spatial expression and function of TaDlea3 in Triticum aestivum during developmental stages under drought stress. Plant Science, 252:290-299.
doi: 10.1016/j.plantsci.2016.08.010 URL |
| [5] |
Christian N, Jean D, Kenneth E, Tessa P, Norman H, Fathey S. 2002. Cold-regulated cereal chloroplast late embryogenesis abundant-like proteins. Molecular characterization and functional analyses. Plant Physiology, 129 (3):1368-1381.
doi: 10.1104/pp.001925 pmid: 12114590 |
| [6] | Cuevas-Velazquez C L, Rendon-Luna D F, Covarrubias A A. 2014. Dissecting the cryoprotection mechanisms for dehydrins. Frontiers in Plant Science, 5:1-6. |
| [7] | Fuxreiter M, Simon I, Friedrich P, Tompa P. 2004. Preformed structural elements feature in partner recognition by intrinsically unstructured proteins. Journal of Molecμlar Biology, 338 (5):1015-1026. |
| [8] |
Gao T, Mo Y X, Huang H Y, Yu J M, Wang Y, Wang W D. 2021. Heterologous expression of Camellia sinensis Late Embryogenesis Abundant Protein Gene 1(CsLEA1)confers cold stress tolerance in Escherichia coli and yeast. Horticultural Plant Journal, 7 (1):89-96.
doi: 10.1016/j.hpj.2020.09.005 URL |
| [9] | Ge Shibei, Jiang Xiaochun, Wang Lingyu, Yu Jingquan, Zhou Yanhong. 2020. Recent advances in the role and mechanism of arbuscular mycorrhizainduced improvement of abiotic stress tolerance in horticultural plan. Acta Horticulturae Sinica, 47 (9):1752-1776. (in Chinese) |
| 葛诗蓓, 姜小春, 王羚羽, 喻景权, 周艳虹. 2020. 园艺植物丛枝菌根抗非生物胁迫的作用机制研究进展. 园艺学报, 47 (9):1752-1776. | |
| [10] |
Grelet J, Benamar A, Teyssier E, Avelange-Macherel M H, Grunwald D, Macherel D. 2005. Identification in pea seed mitochondria of a late-embryogenes is abundant protein able to protect enzymes from drying. Plant Physiology, 137 (1):157-167.
doi: 10.1104/pp.104.052480 URL |
| [11] |
Hu Ting-zhang, Yang Jun-nian, Wu Ying-mei, Chen Zai-gang. 2012. Expression of OsLEA19a gene in Escherichia coli and its resistance analysis. Acta Agriculturae Boreali-Sinica, 27 (6):1-4. (in Chinese)
doi: 10.3969/j.issn.1000-7091.2012.06.001 |
|
胡廷章, 杨俊年, 吴应梅, 陈再刚. 2012. 水稻OsLEA19a基因在大肠杆菌中的表达和抗逆性分析. 华北农学报, 27 (6):1-4.
doi: 10.3969/j.issn.1000-7091.2012.06.001 |
|
| [12] | Hundertmark M, Hincha D K. 2008. LEA(late embryogenesis abundant)proteins and their encoding genes in Arabidopsis thaliana. BioMed Central, 9 (1):118. |
| [13] |
Ingram J, Bartels D. 1996. The molecular basis of dehydration tolerance in plants. Annual Review of Plant Physiology and Plant Molecular Biology, 47:377-403.
pmid: 15012294 |
| [14] |
Iturriaga G, Schneider K, Salamini F, Bartels D. 1992. Expression of desiccation-related proteins from the resurrection plant Craterostigma plantagineum in transgenic tobacco. Plant Molecular Biology, 20 (3):555-558.
doi: 10.1007/BF00040614 pmid: 1421157 |
| [15] |
Liu Y, Liang J N, Sun L P, Yang X H, Li D Q. 2016. Group 3 LEA protein,ZmLEA3,is involved in protection from low temperature stress. Frontiers in Plant Science, 7:1011.
doi: 10.3389/fpls.2016.01011 pmid: 27471509 |
| [16] |
Liu Y, Zheng Y Z. 2005. PM2,a group 3 LEA protein from soybean,and its 22-mer repeating region confer salt tolerance in Escherichia coli. Biochemical and Biophysical Research Communications, 331 (1):325-332.
doi: 10.1016/j.bbrc.2005.03.165 URL |
| [17] |
Liu Zi-cheng, Miao Li-li, Wang Jing-yi, Yang De-long, Mao Xin-guo, Jing Rui-lian. 2016. Cloning and characterization of transcription factor TaWRKY35 in wheat(Triticum aestivum). Scientia Agricultura Sinica, 49 (12):2245-2254. (in Chinese)
doi: 10.3864/j.issn.0578-1752.2016.12.001 |
|
刘自成, 苗丽丽, 王景一, 杨德龙, 毛新国, 景蕊莲. 2016. 普通小麦转录因子基因TaWRKY35的克隆及功能分析. 中国农业科学, 49 (12):2245-2254.
doi: 10.3864/j.issn.0578-1752.2016.12.001 |
|
| [18] | Ma Jing, Sun Wen-ting, Wang Jing, Sui Shun-zhao, Li Ming-yang. 2014. Cloning and expression analysis of a late embryogenesis abundant protein gene CpLEA from Chimonanthus praecox. Acta Horticulturae Sinica, 41 (8):1663-1672. (in Chinese) |
| 马婧, 孙文婷, 王晶, 眭顺照, 李名扬. 2014. 蜡梅胚胎晚期丰富蛋白基因CpLEA的克隆及表达分析. 园艺学报, 41 (8):1663-1672. | |
| [19] |
Mariana A, Juriaan R, Timothy J, Jill M, Wilco L, Henk H. 2019. Structural plasticity of intrinsically disordered lea proteins from Xerophyta schlechteri provides protection in vitro and in vivo. Frontiers in Plant Science, 10 (10):1272-1279.
doi: 10.3389/fpls.2019.01272 URL |
| [20] | Sun M Z, Shen Y, Yin K D, Guo Y X, Cai X X, Yang J K, Zhu Y M, Jia B W, Sun X L. 2019. A late embryogenesis abundant protein GsPM30 interacts with a receptor like cytoplasmic kinase GsCBRLK and regulates environmental stress responses. Plant Science:an International Journal of Experimental Plant Biology, 283:70-82. |
| [21] | Tang Xiao-qian, Yu Li-xia, Wu Xiao-lu, Yan Bo. 2010. Research advance in the group three of late-embryogenesis-abundant proteins and genes. Chinese Bulletin of Life Sciences, 22 (6):551-555. (in Chinese) |
| 汤晓倩, 于丽霞, 武晓璐, 鄢波. 2010. 第三组胚胎晚期丰富蛋白lea3基因研究概述. 生命科学, 22 (6):551-555. | |
| [22] |
Wang L J, Li X F, Chen S Y, Liu G S. 2009. Enhanced drought tolerance in transgenic Leymus chinensis plants with constitutively expressed wheat TaLEA3. Biotechnology Letters, 31 (2):313-319.
doi: 10.1007/s10529-008-9864-5 URL |
| [23] |
Wang W G, Li R, Liu B, Li L, Wang S H, Chen F. 2012. Alternatively splicedtranscripts of group 3 late embryogenesis abundant protein from Pogonatherum paniceum confer different abiotic stress tolerance in Escherichia coli. Journal of Plant Physiology, 169 (15):1559-1564.
doi: 10.1016/j.jplph.2012.06.017 URL |
| [24] | Wang X Y, Zhang L S, Zhang Y N, Bai Z Q, Liu H, Zhang D P. 2017. Triticum aestivum WRAB 18 functions in plastids and confers abiotic stress tolerance when overexpressed in Escherichia coli and Nicotiania benthamiana. PLoS ONE, 12 (2):320-340. |
| [25] |
Yu J N, Zhang J S, Shan L, Chen S Y. 2005. Two new group 3 LEA genes of wheat and their functional analysis in yeast. Journal of Integrative Plant Biology, 47 (11):1372-1381.
doi: 10.1111/jipb.2005.47.issue-11 URL |
| [26] | Zhao Bing, Zhang Qi-xiang. 2008. Variation analysis of floral traits in Chinese Chimonanthus praecox germplasm resources. Acta Horticulturae Sinica, 35 (3):383-388. (in Chinese) |
| 赵冰, 张启翔. 2008. 中国蜡梅种质资源花性状的变异分析. 园艺学报, 35 (3):383-388. |
| [1] | WANG Quancheng, WU Jun, LI Lei, SHI Yanxia, XIE Xuewen, LI Baoju, CHAI Ali. Exploration on the Function of Pathogenicity-related Gene CcTLS1 in Corynespora cassiicola From Cucumber [J]. Acta Horticulturae Sinica, 2023, 50(3): 569-582. |
| [2] | JIANG Yu, TU Xunliang, HE Junrong. Analysis of Differential Expression Genes in Leaves of Leaf Color Mutant of Chinese Orchid [J]. Acta Horticulturae Sinica, 2023, 50(2): 371-381. |
| [3] | TIAN Mingkang, XU Zhixiang, LIU Xiuqun, SUI Shunzhao, LI Mingyang, LI Zhineng. Identification of the AP2 Subfamily Transcription Factors in Chimonanthus praecox and the Functional Study of CpAP2-L11 [J]. Acta Horticulturae Sinica, 2023, 50(2): 382-396. |
| [4] | LIANG Jiali, WU Qisong, CHEN Guangquan, ZHANG Rong, XU Chunxiang, FENG Shujie. Identification of the Neopestalotiopsis musae Pathogen of Banana Leaf Spot Disease [J]. Acta Horticulturae Sinica, 2023, 50(2): 410-420. |
| [5] | Patiguli Maimaitituerxun, LUO Qinghong, Gulinisha Kasimu, SHENG Wei, and JIANG Teng. A New Cultivar of Elaeagnus moorcroftii‘Hongling’in Xinjiang [J]. Acta Horticulturae Sinica, 2022, 49(S2): 67-68. |
| [6] | Patiguli Maimaitituerxun, Gulinisha Kasimu, LUO Qinghong, LIU Liyan, LIU Qiaoling, and Reyihan Awutitashi. A New Cultivar of Elaeagnus moorcroftii‘Yafeng’in Xinjiang [J]. Acta Horticulturae Sinica, 2022, 49(S2): 69-70. |
| [7] | LIN Chunsong, XU Suxia, HUANG Qingyun, ZHANG Xueqin, and ZHANG Wenhui. A New Bougainvillea Cultivar‘Yazhizhilian’with Connateflowers [J]. Acta Horticulturae Sinica, 2022, 49(S2): 237-238. |
| [8] | ZHENG Yiwei, WO Kejun, JIANG Baoxin, ZHOU Ruoyi, and XIE Xiaohong, . A New Azalea Cultivar‘Yongzidie’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 241-242. |
| [9] | PENG Yufu, OUYANG Xueling, YE Chuan, CHEN Hualing, PENG Yong, WANG Guohang, KONG Lingpu, LE Jun, and PENG Huohui, . A New Acer cordatum Cultivar‘Huoyan’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 269-270. |
| [10] | ZHANG Chunying, XIA Xi, SU Ming, ZHANG Jie, and GONG Rui. A New Azalea Cultivar‘Yanzhi’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 167-168. |
| [11] | BI Sisheng, ZHANG Lanying, WANG Hongyong, WANG Zhenmeng, MA Bingyao, GAO Wei, and LIU Guimin, . A New Northern Red Oak Cultivar‘Golden Prince' [J]. Acta Horticulturae Sinica, 2022, 49(S1): 179-180. |
| [12] | JIANG Yajun, CHEN Jiajia, TAN Bin, ZHENG Xianbo, WANG Wei, ZHANG Langlang, CHENG Jun, FENG Jiancan. Function Exploration of PpIDD11 in Regulating Peach Flower Development [J]. Acta Horticulturae Sinica, 2022, 49(9): 1841-1852. |
| [13] | SONG Xingrong, YUAN Puying, HE Xiangda. A New Chimonanthus praecox Cultivar‘Bianzaosu’ [J]. Acta Horticulturae Sinica, 2022, 49(9): 2059-2060. |
| [14] | GUO Yan, ZHANG Shuhang, LI Ying, ZHANG Xinfang, WANG Yingjie, WANG Guangpeng. Diversity Analysis of Leaves Phenotypic Traits of Yanshan Chestnut [J]. Acta Horticulturae Sinica, 2022, 49(8): 1673-1688. |
| [15] | CHEN Lilang, YANG Tianzhang, CAI Ruping, LIN Xiaoman, DENG Nankang, CHE Haiyan, LIN Yating, KONG Xiangyi. Molecular Detection and Identification of Viruses from Passiflora edulis in Hainan [J]. Acta Horticulturae Sinica, 2022, 49(8): 1785-1794. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd