
Acta Horticulturae Sinica ›› 2022, Vol. 49 ›› Issue (11): 2377-2387.doi: 10.16420/j.issn.0513-353x.2021-0524
• Research Papars • Previous Articles Next Articles
CHEN Sijia, WANG Huan, LI Ruirui, WANG Zhuoyi, LUO Jing( ), WANG Caiyun
), WANG Caiyun
Received:2022-03-09
Revised:2022-05-09
Online:2022-11-25
Published:2022-11-25
Contact:
LUO Jing
E-mail:ljcau@mail.hzau.edu.cn
CLC Number:
CHEN Sijia, WANG Huan, LI Ruirui, WANG Zhuoyi, LUO Jing, WANG Caiyun. Characterization of CmMYC2 in Formation of Green Color in Ray Florets of Chrysanthemum[J]. Acta Horticulturae Sinica, 2022, 49(11): 2377-2387.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2021-0524
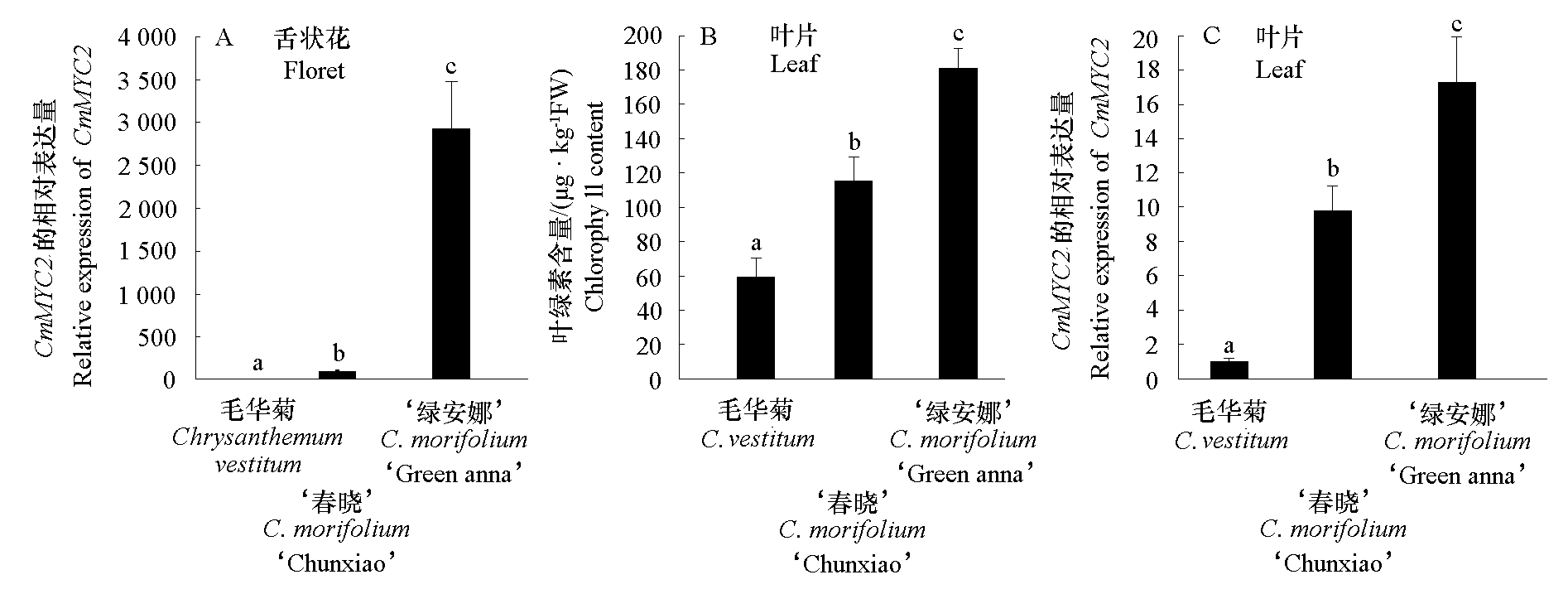
Fig. 1 Expression analysis of CmMYC2 and content of chlorophyll in Chrysanthemum vestitum and C. morifolium‘Chunxiao’and‘Green anna’ Expression of CmMYC2 in ray florets in C. vestitum and C. morifolium ‘Chunxiao’and‘Green anna’(A);Content of chlorophyll(B)and expression of CmMYC2(C)in leaves of C. vestitum and C. morifolium‘Chunxiao’and‘Green anna’.

Fig. 2 Effects of ABA treatment on Chrysanthemum morifolium‘Lükou’phenotype(A,B),chlorophyll content(C),CmMYC2 and chlorophyll synthesis related gene expression(D)

Fig. 3 Phenotype(A),chlorophyll content(B)and relative expression of CmMYC2(C)in CmMYC2 overexpression and Super1300 control samples of Chrysanthemum morifolium‘Lükou’
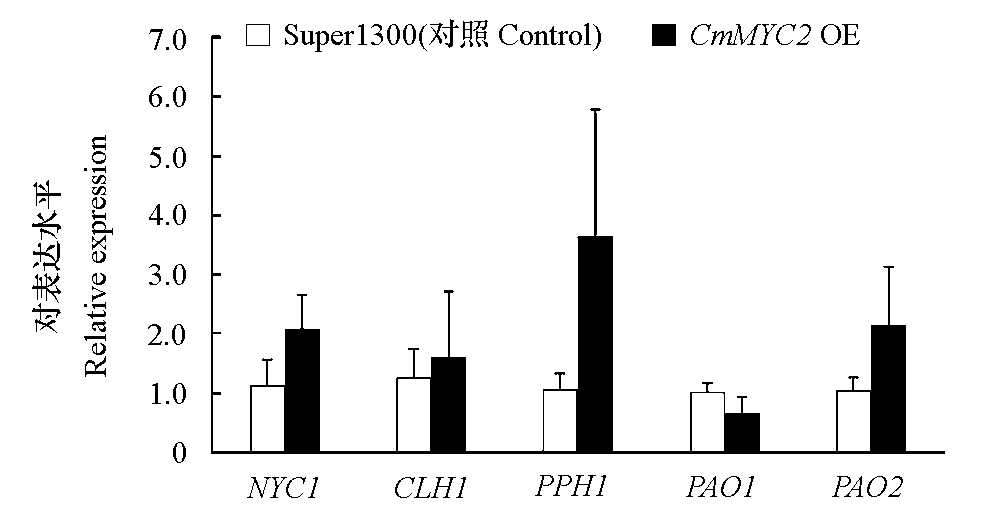
Fig. 5 Analysis of chlorophyll cycling- and degradation-related gene expression in CmMYC2 overexpression and Super1300 control samples of Chrysanthemum morifolium‘Lükou’
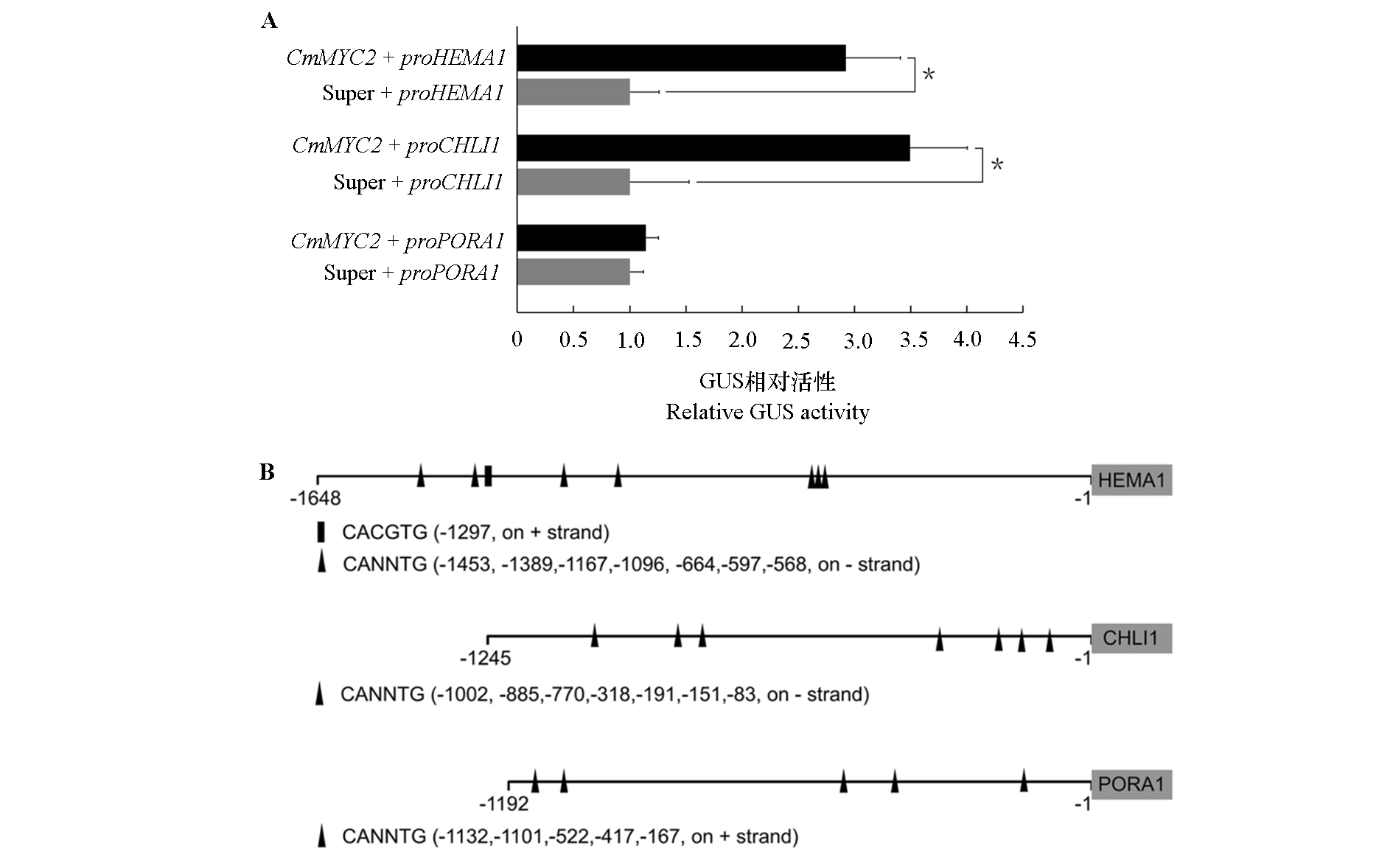
Fig. 6 Regulation of promoter activity of chlorophyll synthesis gene by CmMYC2 A. Transient transactivation assays of HEMA1,CHLI1 and PORA1 promoters by CmMYC2;B. MYC binding sites in the promoters of HEMA1,CHLI1 and PORA1. Rectangles and triangles represent the binding sites on the + or - chains,respectively.
| [1] |
Chen L, Xiang S, Chen Y, Li D, Yu D. 2017. Arabidopsis WRKY 45 interacts with the DELLA protein RGL1 to positively regulate age-triggered leaf senescence. Molecular Plant, 10 (9):1174-1189.
doi: 10.1016/j.molp.2017.07.008 URL |
| [2] |
Dong W, Li M, Li Z, Li S, Zhu Y, Wang Z. 2020. Transcriptome analysis of the molecular mechanism of Chrysanthemum flower color change under short-day photoperiods. Plant Physiology and Biochemistry, 146:315-328.
doi: S0981-9428(19)30492-9 pmid: 31785518 |
| [3] |
Fan Z Q, Tan X L, Shan W, Kuang J F, Lu W J, Chen J Y. 2017. BrWRKY65,a WRKY transcription factor,is involved in regulating three leaf senescence-associated genes in Chinese flowering cabbage. International Journal of Molecular Sciences, 18 (6):1228.
doi: 10.3390/ijms18061228 URL |
| [4] |
Fu H, Zeng T, Zhao Y, Luo T, Deng H, Meng C, Luo J, Wang C. 2021. Identification of chlorophyll metabolism- and photosynthesis-related genes regulating green flower color in chrysanthemum by integrative transcriptome and weighted correlation network analyses. Genes, 12 (3):449.
doi: 10.3390/genes12030449 URL |
| [5] | Hong Yan, Bai Xinxiang, Sun Wei, Jia Fengwei,Dai Silan. 2012. Quantitative classification of flower color phenotypes of chrysanthemum cultivars. Acta horticulturae Sinica, 39 (7):1330-1340. (in Chinese) |
| 洪艳, 白新祥, 孙卫, 贾锋炜, 戴思兰. 2012. 菊花品种花色表型数量分类研究. 园艺学报, 39 (7):1330-1340. | |
| [6] |
Hu B, Lai B, Wang D, Li J Q, Chen L H, Qin Y Q, Wang H C, Qin Y H, Hu G B, Zhao J T. 2019. Three LcABFs are involved in the regulation of chlorophyll degradation and anthocyanin biosynthesis during fruit ripening in Litchi chinensis. Plant and Cell Physiology, 60 (2):448-461.
doi: 10.1093/pcp/pcy219 URL |
| [7] |
Job N, Datta S. 2021. PIF3/HY 5 module regulates BBX11 to suppress protochlorophyllide levels in dark and promote photomorphogenesis in light. New Phytologist, 230 (1):190-204.
doi: 10.1111/nph.17149 URL |
| [8] |
Kalaji H M, Dąbrowski P, Cetner M D, Samborska I A, Łukasik I, Brestic M, Zivcak M, Tomasz H, Mojski J, Kociel H, Panchal B M. 2017. A comparison between different chlorophyll content meters under nutrient deficiency conditions. Journal of Plant Nutrition, 40 (7):1024-1034.
doi: 10.1080/01904167.2016.1263323 URL |
| [9] |
Kazan K, Manners J M. 2013. MYC2:the master in action. Molecular Plant, 6 (3):686-703.
doi: 10.1093/mp/sss128 URL |
| [10] |
Kishimoto S, Maoka T, Nakayama M, Ohmiya A. 2004. Carotenoid composition in petals of chrysanthemum[Dendranthema grandiflorum (Ramat.)Kitamura]. Phytochemistry, 65 (20):2781-2787.
pmid: 15474564 |
| [11] | Li he-sheng. 2000. Principles and techniques of plant physiological biochemical experiment. Beijing: Higher Education Press. (in Chinese) |
| 李合生. 2000. 植物生理生化实验原理和技术. 北京: 高等教育出版社. | |
| [12] |
Li S, Xie X, Liu S, Chen K, Yin X. 2019. Auto- and mutual-regulation between two CitERFs contribute to ethylene-induced citrus fruit degreening. Food Chemistry, 299:125163.
doi: 10.1016/j.foodchem.2019.125163 URL |
| [13] |
Li W, Wang L, He Z, Lu Z, Cui J, Xu N, Jin B, Wang L. 2020. Physiological and transcriptomic changes during autumn coloration and senescence in Ginkgo biloba leaves. Horticultural Plant Journal, 6 (6):396-408.
doi: 10.1016/j.hpj.2020.11.002 URL |
| [14] |
Li X, Duan X, Jiang H, Sun Y, Tang Y, Yuan Z, Guo J, Liang W, Chen L, Yin J, Ma H, Wang J, Zhang D. 2006. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiology, 141:1167-1184.
doi: 10.1104/pp.106.080580 URL |
| [15] |
Liu X, Li Y, Zhong S. 2017. Interplay between light and plant hormones in the control of Arabidopsis seedling chlorophyll biosynthesis. Frontiers in Plant Science, 8:1433.
doi: 10.3389/fpls.2017.01433 URL |
| [16] |
Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods, 25 (4):402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [17] |
Luo J, Wang H, Chen S, Ren S, Fu H, Li R, Wang C. 2021. CmNAC 73 mediates the formation of green color in chrysanthemum flowers by directly activating the expression of chlorophyll biosynthesis genes HEMA1 and CRD1. Genes, 12 (5):704.
doi: 10.3390/genes12050704 URL |
| [18] |
Mathew I E, Agarwal P. 2018. May the fittest protein evolve:favoring the plant-specific origin and expansion of NAC transcription factors. BioEssays, 40 (8):1800018.
doi: 10.1002/bies.201800018 URL |
| [19] |
Ohmiya A, Hirashima M, Yagi M, Tanase K, Yamamizo C. 2014. Identification of genes associated with chlorophyll accumulation in flower petals. PLoS ONE, 9:e113738.
doi: 10.1371/journal.pone.0113738 URL |
| [20] |
Ohmiya A, Oda-Yamamizo C, Kishimoto S. 2019. Overexpression of CONSTANS-like 16 enhances chlorophyll accumulation in petunia corollas. Plant Science, 280:90-96.
doi: S0168-9452(18)30932-4 pmid: 30824032 |
| [21] |
Ohmiya A, Sasaki K, Nashima K, Oda-Yamamizo C, Hirashima M, Sumitomo K. 2017. Transcriptome analysis in petals and leaves of chrysanthemums with different chlorophyll levels. BMC Plant Biology, 17 (1):1-15.
doi: 10.1186/s12870-016-0951-9 URL |
| [22] | Sakuraba Y, Han S H, Lee S H, Hörtensteiner S, Paek N C. 2016. Arabidopsis NAC016 promotes chlorophyll breakdown by directly upregulating STAYGREEN1 transcription. Plant cell Report, 35 (1):155-166. |
| [23] | Shi dianyi, Liu Zhongxiang, Jin Weiwei. 2009. Chlorophyll synthesis,catabolism and signal regulation in plants. Genetics, 31 (7):698-704. (in Chinese) |
| 史典义, 刘忠香, 金危危. 2009. 植物叶绿素合成,分解代谢及信号调控. 遗传, 31 (7):698-704. | |
| [24] | Tanaka Y, Sasaki N. 2008. Plant pigments for coloration. Plant Journal, 135 (1):733-749. |
| [25] |
Toledo-Ortiz G, Huq E, Quail P H. 2003. The Arabidopsis basic/helix-loop-helix transcription factor family. The Plant Cell, 15 (8):1749-1770.
doi: 10.1105/tpc.013839 URL |
| [26] |
Wang C, Dai S, Zhang Z L, Lao W, Wang R, Meng X, Zhou X. 2021a. Ethylene and salicylic acid synergistically accelerate leaf senescence in Arabidopsis. Journal of Integrative Plant Biology, 63 (5):828-833.
doi: 10.1111/jipb.13075 URL |
| [27] |
Wang H L, Zhang Y, Wang T, Yang Q, Yang Y L, Li Z, Li B S, Wen X, Li W Y, Yin W L, Xia X, Guo H, Li Z. 2021b. An alternative splicing variant of PtRD 26 delays leaf senescence by regulating multiple NAC transcription factors in populus. The Plant Cell, 33 (5):1594-1614.
doi: 10.1093/plcell/koab046 URL |
| [28] |
Woo H R, Kim H J, Lim P O, Nam H G. 2019. Leaf senescence:Systems and dynamics aspects. Annual Review of Plant Biology, 70:347-376.
doi: 10.1146/annurev-arplant-050718-095859 URL |
| [29] | Xu L F, Yang P P, Feng Y Y, Xu H, Cao Y W, Tang Y C, Yuan S X, Liu X Y, Ming J. 2017. Spatiotemporal transcriptome analysis provides insights into bicolor tepal development in Lilium“Tiny Padhye”. Frontiers in Plant Science, 8:398. |
| [30] |
Yan Y, Li C, Dong X, Li H, Zhang D, Zhou Y, Jiang B, Peng J, Qin X, Cheng J, Wang X, Song P, Qi L, Zheng Y, Li B, Terzaghi W, Yang S, Guo Y, Li J. 2020. MYB 30 is a key negative regulator of Arabidopsis photomorphogenic development that promotes PIF4 and PIF5 protein accumulation in the light. The Plant Cell, 32 (7):2196-2215.
doi: 10.1105/tpc.19.00645 pmid: 32371543 |
| [31] |
Yu X, Xu Y, Yan S. 2021a. Salicylic acid and ethylene coordinately promote leaf senescence. Journal of Integrative Plant Biology, 63 (5):823-827.
doi: 10.1111/jipb.13074 URL |
| [32] |
Yu J Q, Gu K D, Sun C H, Zhang Q Y, Wang J H, Ma F F, You C X, Hu D G, Hao Y J. 2021b. The apple bHLH transcription factor MdbHLH 3 functions in determining the fruit carbohydrates and malate. Plant Biotechnology Journal, 19 (2):285-299.
doi: 10.1111/pbi.13461 URL |
| [33] | Zhang Lijun, Dai Silan. 2009. Research Advance on Germplasm Resources of Chrysanthemum × morifolium. Bulletin of Botany, 44 (5):526-535. (in Chinese) |
| 张莉俊, 戴思兰. 2009. 菊花种质资源研究进展. 植物学报, 44 (5):526-535. | |
| [34] |
Zhang W, Peng K, Cui F, Wang D, Zhao J, Zhang Y, Yu N, Wang Y, Zeng D, Wang Y, Cheng Z, Zhang K. 2021. Cytokinin oxidase/dehydrogenase OsCKX11 coordinates source and sink relationship in rice by simultaneous regulation of leaf senescence and grain number. Plant biotechnology journal, 19 (2):335-350.
doi: 10.1111/pbi.13467 pmid: 33448635 |
| [35] |
Zhang Y, Li Y, Hassan M J, Li Z, Peng Y. 2020. Indole-3-acetic acid improves drought tolerance of white clover via activating auxin, abscisic acid and jasmonic acid related genes and inhibiting senescence genes. BMC Plant Biology, 20:1-12.
doi: 10.1186/s12870-019-2170-7 URL |
| [36] | Zhao Li. 2011 Influence of bHLH homologous gene expression on flower coloration of chrysanthemum[M. D. Dissertation]. Beijing: Beijing Forestry University. (in Chinese) |
| 赵莉. 2011. bHLH同源基因的表达对菊花花色的影响[硕士论文]. 北京: 北京林业大学. | |
| [37] | Zhu Anchao. 2014. Investigation on mechanism of green disk florets of spray cut chrysanthemum[M. D. Dissertation]. Nanjing: Nanjing Agricultural University. (in Chinese) |
| 朱安超. 2014. “绿心”切花小菊管状花绿色形成机理研究[硕士论文]. 南京: 南京农业大学. | |
| [38] |
Zhu X, Chen J, Xie Z, Gao J, Ren G, Gao S, Zhou X, Kuai B. 2015. Jasmonic acid promotes degreening via MYC 2/3/4‐and ANAC 019/055/072‐mediated regulation of major chlorophyll catabolic genes. The Plant Journal, 84 (3):597-610.
doi: 10.1111/tpj.13030 URL |
| [39] |
Zhu Z, Li G, Yan C, Liu L, Zhang Q, Han Z, Li B. 2019. DRL1,encoding a NAC transcription factor,is involved in leaf senescence in grapevine. International Journal of Molecular Sciences, 20 (11):2678.
doi: 10.3390/ijms20112678 URL |
| [1] | YE Zimao, SHEN Wanxia, LIU Mengyu, WANG Tong, ZHANG Xiaonan, YU Xin, LIU Xiaofeng, and ZHAO Xiaochun, . Effect of R2R3-MYB Transcription Factor CitMYB21 on Flavonoids Biosynthesis in Citrus [J]. Acta Horticulturae Sinica, 2023, 50(2): 250-264. |
| [2] | SONG Yanhong, CHEN Yaduo, ZHANG Xiaoyu, SONG Pan, LIU Lifeng, LI Gang, ZHAO Xia, and ZHOU Houcheng, . The Transcription Factor FvbHLH130 Activates Flowering in Fragaria vesca [J]. Acta Horticulturae Sinica, 2023, 50(2): 295-306. |
| [3] | HAN Rui, ZHONG Xionghui, CHEN Denghui, CUI Jian, YUE Xiangqing, XIE Jianming, and KANG Jungen, . Cloning and Functional Analysis of BobHLH34 Gene in Cabbage that Interacts with XopR from Xanthomonas [J]. Acta Horticulturae Sinica, 2023, 50(2): 319-330. |
| [4] | TIAN Mingkang, XU Zhixiang, LIU Xiuqun, SUI Shunzhao, LI Mingyang, and LI Zhineng, . Identification of the AP2 Subfamily Transcription Factors in Chimonanthus praecox and the Functional Study of CpAP2-L11 [J]. Acta Horticulturae Sinica, 2023, 50(2): 382-396. |
| [5] | LIN Haijiao, LIANG Yuchen, LI Ling, MA Jun, ZHANG Lu, LAN Zhenying, YUAN Zening. Exploration and Regulation Network Analysis of CBF Pathway Related Cold Tolerance Genes in Lavandula angustifolia [J]. Acta Horticulturae Sinica, 2023, 50(1): 131-144. |
| [6] | TAN Shanshan, QIU Liang, DUAN Aoqi, SHU Sheng, ZENG Xiaoping, ZHU Weimin, JIA Min, YAN Jun, LIU Yanhua, LIU Hui, XIONG Aisheng. Effects of Different Concentrations of Sodium Hypochlorite on Seeds Germination and Seedlings Quality of Celery [J]. Acta Horticulturae Sinica, 2022, 49(9): 1907-1921. |
| [7] | JIA Xin, ZENG Zhen, CHEN Yue, FENG Hui, LÜ Yingmin, ZHAO Shiwei. Cloning and Expression Analysis of RcDREB2A Gene in Rosa chinensis‘Old Blush’ [J]. Acta Horticulturae Sinica, 2022, 49(9): 1945-1956. |
| [8] | XU Haifeng, WANG Zhongtang, CHEN Xin, LIU Zhiguo, WANG Lihu, LIU Ping, LIU Mengjun, ZHANG Qiong. The Analyses of Target Metabolomics in Flavonoid and Its Potential MYB Regulation Factors During Coloring Period of Winter Jujube [J]. Acta Horticulturae Sinica, 2022, 49(8): 1761-1771. |
| [9] | QIAN Jieyu, JIANG Lingli, ZHENG Gang, CHEN Jiahong, LAI Wuhao, XU Menghan, FU Jianxin, ZHANG Chao. Identification and Expression Analysis of MYB Transcription Factors Regulating the Anthocyanin Biosynthesis in Zinnia elegans and Function Research of ZeMYB9 [J]. Acta Horticulturae Sinica, 2022, 49(7): 1505-1518. |
| [10] | CHEN Daozong, LIU Yi, SHEN Wenjie, ZHU Bo, TAN Chen. Identification and Analysis of PAP1/2 Homologous Genes in Brassica rapa,B. oleracea and B. napus [J]. Acta Horticulturae Sinica, 2022, 49(6): 1301-1312. |
| [11] | WANG Yan, SUN Zheng, FENG Shan, YUAN Xinyi, ZHONG Linlin, ZENG Yunliu, FU Xiaopeng, CHENG Yunjiang, Bao Manzhu, ZHANG Fan. The Negative Regulation of DcERF-1 on Senescence of Cut Carnation [J]. Acta Horticulturae Sinica, 2022, 49(6): 1313-1326. |
| [12] | SHEN Zhiguo, ZHANG Lin, YUAN Deyi, CHENG Jianming. Research Progress on Flower Color and New Red Flower Resource of Wintersweets(Chimonanthus praecox) [J]. Acta Horticulturae Sinica, 2022, 49(4): 924-934. |
| [13] | WANG Jing, XU Leifeng, WANG Ling, QI Xianyu, SONG Meng, CAO Yuwei, HE Guoren, TANG Yuchao, YANG Panpan, MING Jun. The Numerical Classification of Flower Color Phenotype in Lily [J]. Acta Horticulturae Sinica, 2022, 49(3): 571-580. |
| [14] | ZHOU Lin, ZOU Hongzhu, HAN Lulu, JIA Yinghua, WANG Yan. Research Progress on the Role of Glycosyltransferases in Color Formation of Petals [J]. Acta Horticulturae Sinica, 2022, 49(3): 687-700. |
| [15] | LI Chunhong, WANG Kaituo, LEI Changyi, XU Feng, JI Nana, JIANG Yongbo. Identification of TGA Gene Family in Peach and Analysis of Expression Mode Involved in a BABA-Induced Disease Resistance [J]. Acta Horticulturae Sinica, 2022, 49(2): 265-280. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd