
Acta Horticulturae Sinica ›› 2022, Vol. 49 ›› Issue (11): 2313-2324.doi: 10.16420/j.issn.0513-353x.2021-0863
• Research Papars • Previous Articles Next Articles
NIE Xinghua1,2, LI Yiran2, TIAN Shoule3, WANG Xuefeng4, SU Shuchai1, CAO Qingqin2, XING Yu2,*( ), QIN Ling2,*(
), QIN Ling2,*( )
)
Received:2022-04-26
Revised:2022-07-28
Online:2022-11-25
Published:2022-11-25
Contact:
XING Yu,QIN Ling
E-mail:xingyu@bua.edu.cn;qinlingbac@126.com
CLC Number:
NIE Xinghua, LI Yiran, TIAN Shoule, WANG Xuefeng, SU Shuchai, CAO Qingqin, XING Yu, QIN Ling. Construction of DNA Fingerprint Map and Analysis of Genetic Diversity for Chinese Chestnut Cultivars(Lines)[J]. Acta Horticulturae Sinica, 2022, 49(11): 2313-2324.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2021-0863
| 编号 Number | 分子标记 Marker ID | 基因库ID GenBank ID | 正向引物序列(5′-3′) Forward primer | 反向引物序列(5′-3′) Reverse primer | 观察的重复序列 Observed-Motif |
|---|---|---|---|---|---|
| 1 | CmSI0396 | 290474606 | AACTCCCACCACTCACATCC | TTTCGGACCATCCAGAACTC | CACACC |
| 2 | CmSI0509 | 290474647 | CAAGTCCGATCCTTCCTCTG | AGCTGGGTTTTGAGTAGCGA | ACA |
| 3 | CmSI0561 | 290474702 | CGTATAGGGTGGAAACGGAA | GGACAAGCAAATCACGGAAT | TCG |
| 4 | CmSI0614 | 290476556 | TTGTGGTGAAGCTGACATCG | GGGTACTACCACAACATGCAG | GTT |
| 5 | CmSI0617 | 290475843 | GCCTCAGCTGGTAACGAAGA | TCACATGGGCTTGTTGTAGC | GA |
| 6 | CmSI0658 | 290474781 | AAAACGGTTTGTGGTGAAGC | GCCAACCAGTCAAGGGTACT | GTT |
| 7 | CmSI0702 | 290474818 | GAAACACACCAGAGAGATGCAG | TTTTATACAGAGACATACTATCCTACACAG | TC |
| 8 | CmSI0735 | 290474842 | ACGCCTTCAGTTGCTGTTTC | CAACGGTCTTACCCTTTGGA | AGAA |
| 9 | CmSI0742 | 290474850 | GACGCTCCTCAGCTTTTGAC | TGCCGGTCAATTCTTCTTCT | AG |
| 10 | CmSI0800 | 290474899 | TTATGGCAACCCTCCTGTTT | CTGAAATGATCGATGCTGCT | TC |
| 11 | CmSI0853 | 290474950 | GGAGGAGGAGGAGCTCATTG | CCTTGGAGAGCTGCCAGTAG | TCT |
| 12 | CmSI0871 | 290474967 | AGGGGGTGGAAGAACCTATG | AGATTGCAAGTGGGGAATTG | TCT |
| 13 | CmSI0881 | 290476058 | TCCGACAAGCTACCGAGTCT | CCTAAATCAATCTTGCACCCATA | GA |
| 14 | CmSI0883 | 290476060 | CAGCATCAGCACTCGTTCA | GGGATTGAGAGGATGAAGCA | AGC |
| 15 | CmSI0922 | 290475011 | AATCTGAACCCCTCCGATCT | ACCAACAACATGTGCCAAAA | TTG |
| 16 | CmSI0930 | 290475019 | CCATTTAGCATGCATAGTCATACC | GCAAGGATGTAGGTCGAATCA | ATAC |
| 17 | CmSI0938 | 290475031 | CACGATTCCCACGATTCTTA | CGTCCAACTCCGTACTCTCC | GAC |
| 18 | CmSI0404 | 290474610 | GTGCTACGACTACCGCTGCT | AGAATAACCTCGCGGTGAGA | ACA |
| 19 | CmSI0747 | 290474854 | CATTCCAACAATGCACTCTCA | CGAGTTGGAGTCACCGAAGT | CTT |
| 20 | CmSI0809 | 290474901 | GAGCGAGTCGAAGAAGGAAG | ATCTGCTTCGGCACCATCT | GGT |
| 21 | CmSI0933 | 290475025 | AGGCCTCTTCTCCCTTGTGT | TCCTGTTCCTATGCTGCTCA | AGA |
Table 1 The information of 21 pairs of SSR primers
| 编号 Number | 分子标记 Marker ID | 基因库ID GenBank ID | 正向引物序列(5′-3′) Forward primer | 反向引物序列(5′-3′) Reverse primer | 观察的重复序列 Observed-Motif |
|---|---|---|---|---|---|
| 1 | CmSI0396 | 290474606 | AACTCCCACCACTCACATCC | TTTCGGACCATCCAGAACTC | CACACC |
| 2 | CmSI0509 | 290474647 | CAAGTCCGATCCTTCCTCTG | AGCTGGGTTTTGAGTAGCGA | ACA |
| 3 | CmSI0561 | 290474702 | CGTATAGGGTGGAAACGGAA | GGACAAGCAAATCACGGAAT | TCG |
| 4 | CmSI0614 | 290476556 | TTGTGGTGAAGCTGACATCG | GGGTACTACCACAACATGCAG | GTT |
| 5 | CmSI0617 | 290475843 | GCCTCAGCTGGTAACGAAGA | TCACATGGGCTTGTTGTAGC | GA |
| 6 | CmSI0658 | 290474781 | AAAACGGTTTGTGGTGAAGC | GCCAACCAGTCAAGGGTACT | GTT |
| 7 | CmSI0702 | 290474818 | GAAACACACCAGAGAGATGCAG | TTTTATACAGAGACATACTATCCTACACAG | TC |
| 8 | CmSI0735 | 290474842 | ACGCCTTCAGTTGCTGTTTC | CAACGGTCTTACCCTTTGGA | AGAA |
| 9 | CmSI0742 | 290474850 | GACGCTCCTCAGCTTTTGAC | TGCCGGTCAATTCTTCTTCT | AG |
| 10 | CmSI0800 | 290474899 | TTATGGCAACCCTCCTGTTT | CTGAAATGATCGATGCTGCT | TC |
| 11 | CmSI0853 | 290474950 | GGAGGAGGAGGAGCTCATTG | CCTTGGAGAGCTGCCAGTAG | TCT |
| 12 | CmSI0871 | 290474967 | AGGGGGTGGAAGAACCTATG | AGATTGCAAGTGGGGAATTG | TCT |
| 13 | CmSI0881 | 290476058 | TCCGACAAGCTACCGAGTCT | CCTAAATCAATCTTGCACCCATA | GA |
| 14 | CmSI0883 | 290476060 | CAGCATCAGCACTCGTTCA | GGGATTGAGAGGATGAAGCA | AGC |
| 15 | CmSI0922 | 290475011 | AATCTGAACCCCTCCGATCT | ACCAACAACATGTGCCAAAA | TTG |
| 16 | CmSI0930 | 290475019 | CCATTTAGCATGCATAGTCATACC | GCAAGGATGTAGGTCGAATCA | ATAC |
| 17 | CmSI0938 | 290475031 | CACGATTCCCACGATTCTTA | CGTCCAACTCCGTACTCTCC | GAC |
| 18 | CmSI0404 | 290474610 | GTGCTACGACTACCGCTGCT | AGAATAACCTCGCGGTGAGA | ACA |
| 19 | CmSI0747 | 290474854 | CATTCCAACAATGCACTCTCA | CGAGTTGGAGTCACCGAAGT | CTT |
| 20 | CmSI0809 | 290474901 | GAGCGAGTCGAAGAAGGAAG | ATCTGCTTCGGCACCATCT | GGT |
| 21 | CmSI0933 | 290475025 | AGGCCTCTTCTCCCTTGTGT | TCCTGTTCCTATGCTGCTCA | AGA |
| 分子标记 Marker ID | 主要位点 频率MAF | 基因型数Ng | 位点数 Na | Shannon多样性指数I | 观察杂合度Ho | 期望杂合度He | 基因流 Nm | 多态信息 含量 PIC | 身份概率 PI | 随机身份 概率 PIsibs |
|---|---|---|---|---|---|---|---|---|---|---|
| CmSI0396 | 0.568 | 9 | 4 | 0.989 | 0.589 | 0.567 | 7.678 | 0.492 | 0.261 | 0.532 |
| CmSI0509 | 0.477 | 13 | 6 | 1.179 | 0.542 | 0.646 | 4.987 | 0.583 | 0.188 | 0.474 |
| CmSI0561 | 0.509 | 32 | 14 | 1.602 | 0.630 | 0.694 | 3.793 | 0.673 | 0.118 | 0.433 |
| CmSI0617 | 0.325 | 30 | 9 | 1.713 | 0.714 | 0.778 | 3.482 | 0.744 | 0.081 | 0.381 |
| CmSI0658 | 0.249 | 22 | 7 | 1.703 | 0.752 | 0.802 | 5.099 | 0.774 | 0.069 | 0.366 |
| CmSI0702 | 0.412 | 26 | 11 | 1.409 | 0.647 | 0.702 | 3.202 | 0.655 | 0.140 | 0.434 |
| CmSI0735 | 0.403 | 27 | 10 | 1.604 | 0.557 | 0.732 | 9.646 | 0.697 | 0.109 | 0.411 |
| CmSI0742 | 0.422 | 20 | 13 | 1.346 | 0.627 | 0.689 | 6.857 | 0.633 | 0.151 | 0.443 |
| CmSI0800 | 0.326 | 58 | 22 | 2.020 | 0.633 | 0.802 | 8.051 | 0.777 | 0.063 | 0.365 |
| CmSI0809 | 0.270 | 24 | 10 | 1.824 | 0.624 | 0.809 | 2.930 | 0.762 | 0.063 | 0.362 |
| CmSI0853 | 0.301 | 26 | 14 | 1.740 | 0.749 | 0.792 | 7.355 | 0.762 | 0.073 | 0.372 |
| CmSI0871 | 0.597 | 15 | 11 | 0.920 | 0.522 | 0.527 | 11.653 | 0.448 | 0.306 | 0.563 |
| CmSI0883 | 0.278 | 33 | 13 | 1.787 | 0.636 | 0.798 | 4.404 | 0.770 | 0.070 | 0.368 |
| CmSI0933 | 0.646 | 10 | 7 | 0.936 | 0.493 | 0.514 | 10.042 | 0.459 | 0.294 | 0.567 |
| CmSI0938 | 0.785 | 14 | 7 | 0.755 | 0.204 | 0.364 | 10.284 | 0.332 | 0.432 | 0.676 |
| CmSI0922 | 0.254 | 38 | 12 | 1.944 | 0.738 | 0.833 | 7.656 | 0.811 | 0.049 | 0.346 |
| CmSI0930 | 0.419 | 51 | 20 | 1.885 | 0.697 | 0.763 | 9.199 | 0.741 | 0.080 | 0.389 |
| CmSI0881 | 0.324 | 24 | 8 | 1.760 | 0.493 | 0.798 | 6.470 | 0.756 | 0.068 | 0.368 |
| CmSI0614 | 0.212 | 29 | 10 | 1.857 | 0.761 | 0.826 | 6.203 | 0.800 | 0.054 | 0.351 |
| CmSI0404 | 0.839 | 6 | 4 | 0.781 | 0.224 | 0.388 | 2.285 | 0.254 | 0.400 | 0.656 |
| CmSI0747 | 0.549 | 10 | 5 | 1.382 | 0.487 | 0.676 | 7.158 | 0.578 | 0.143 | 0.448 |
| 平均值Mean | 0.436 | 24.619 | 10.333 | 1.483 | 0.587 | 0.690 | 6.592 | 0.643 | 0.153 | 0.443 |
| 组合 Combined | — | — | — | — | — | — | — | — | 4.960 ×10-20 | 2.395×10-8 |
Table 2 The key genetic statistics of 21 SSR markers in 342 Chinese chestnut accessions
| 分子标记 Marker ID | 主要位点 频率MAF | 基因型数Ng | 位点数 Na | Shannon多样性指数I | 观察杂合度Ho | 期望杂合度He | 基因流 Nm | 多态信息 含量 PIC | 身份概率 PI | 随机身份 概率 PIsibs |
|---|---|---|---|---|---|---|---|---|---|---|
| CmSI0396 | 0.568 | 9 | 4 | 0.989 | 0.589 | 0.567 | 7.678 | 0.492 | 0.261 | 0.532 |
| CmSI0509 | 0.477 | 13 | 6 | 1.179 | 0.542 | 0.646 | 4.987 | 0.583 | 0.188 | 0.474 |
| CmSI0561 | 0.509 | 32 | 14 | 1.602 | 0.630 | 0.694 | 3.793 | 0.673 | 0.118 | 0.433 |
| CmSI0617 | 0.325 | 30 | 9 | 1.713 | 0.714 | 0.778 | 3.482 | 0.744 | 0.081 | 0.381 |
| CmSI0658 | 0.249 | 22 | 7 | 1.703 | 0.752 | 0.802 | 5.099 | 0.774 | 0.069 | 0.366 |
| CmSI0702 | 0.412 | 26 | 11 | 1.409 | 0.647 | 0.702 | 3.202 | 0.655 | 0.140 | 0.434 |
| CmSI0735 | 0.403 | 27 | 10 | 1.604 | 0.557 | 0.732 | 9.646 | 0.697 | 0.109 | 0.411 |
| CmSI0742 | 0.422 | 20 | 13 | 1.346 | 0.627 | 0.689 | 6.857 | 0.633 | 0.151 | 0.443 |
| CmSI0800 | 0.326 | 58 | 22 | 2.020 | 0.633 | 0.802 | 8.051 | 0.777 | 0.063 | 0.365 |
| CmSI0809 | 0.270 | 24 | 10 | 1.824 | 0.624 | 0.809 | 2.930 | 0.762 | 0.063 | 0.362 |
| CmSI0853 | 0.301 | 26 | 14 | 1.740 | 0.749 | 0.792 | 7.355 | 0.762 | 0.073 | 0.372 |
| CmSI0871 | 0.597 | 15 | 11 | 0.920 | 0.522 | 0.527 | 11.653 | 0.448 | 0.306 | 0.563 |
| CmSI0883 | 0.278 | 33 | 13 | 1.787 | 0.636 | 0.798 | 4.404 | 0.770 | 0.070 | 0.368 |
| CmSI0933 | 0.646 | 10 | 7 | 0.936 | 0.493 | 0.514 | 10.042 | 0.459 | 0.294 | 0.567 |
| CmSI0938 | 0.785 | 14 | 7 | 0.755 | 0.204 | 0.364 | 10.284 | 0.332 | 0.432 | 0.676 |
| CmSI0922 | 0.254 | 38 | 12 | 1.944 | 0.738 | 0.833 | 7.656 | 0.811 | 0.049 | 0.346 |
| CmSI0930 | 0.419 | 51 | 20 | 1.885 | 0.697 | 0.763 | 9.199 | 0.741 | 0.080 | 0.389 |
| CmSI0881 | 0.324 | 24 | 8 | 1.760 | 0.493 | 0.798 | 6.470 | 0.756 | 0.068 | 0.368 |
| CmSI0614 | 0.212 | 29 | 10 | 1.857 | 0.761 | 0.826 | 6.203 | 0.800 | 0.054 | 0.351 |
| CmSI0404 | 0.839 | 6 | 4 | 0.781 | 0.224 | 0.388 | 2.285 | 0.254 | 0.400 | 0.656 |
| CmSI0747 | 0.549 | 10 | 5 | 1.382 | 0.487 | 0.676 | 7.158 | 0.578 | 0.143 | 0.448 |
| 平均值Mean | 0.436 | 24.619 | 10.333 | 1.483 | 0.587 | 0.690 | 6.592 | 0.643 | 0.153 | 0.443 |
| 组合 Combined | — | — | — | — | — | — | — | — | 4.960 ×10-20 | 2.395×10-8 |
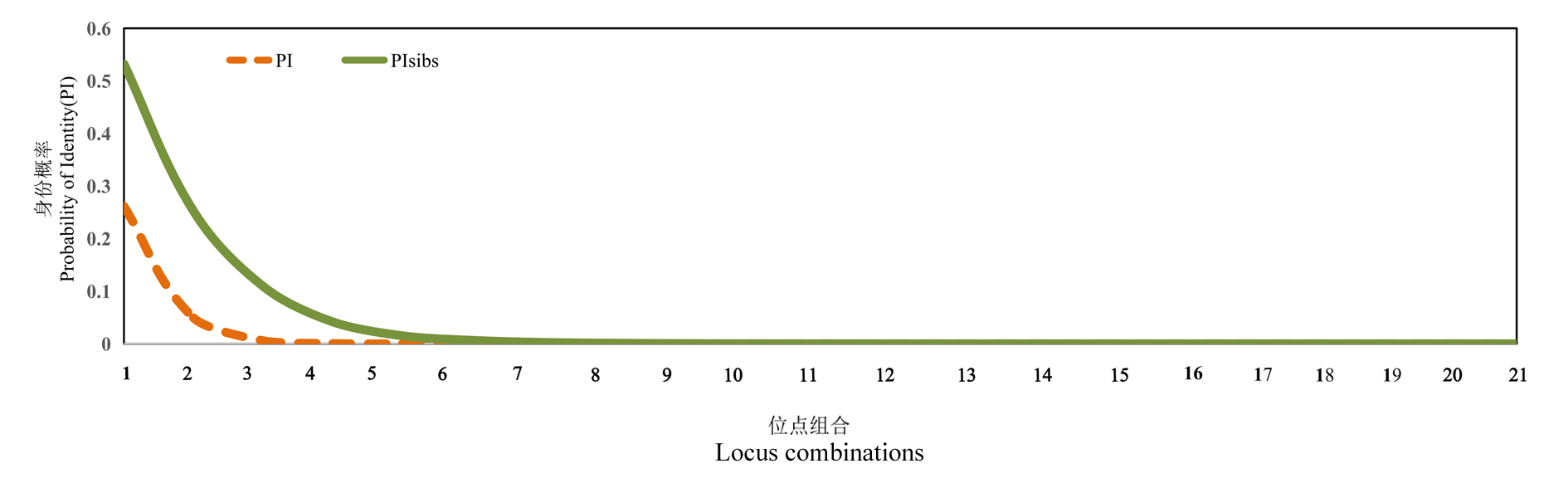
Fig. 1 Evaluation for the fingerprinting power of SSR markers in Chinese chestnut cultivars 1 represents primer 1;2 represents primer 1 + primer 2;3 represents primer 1 + primer 2 + primer 3;......;21 represents all 21 primer combinations. The primer numbers are the same as in Table 1.
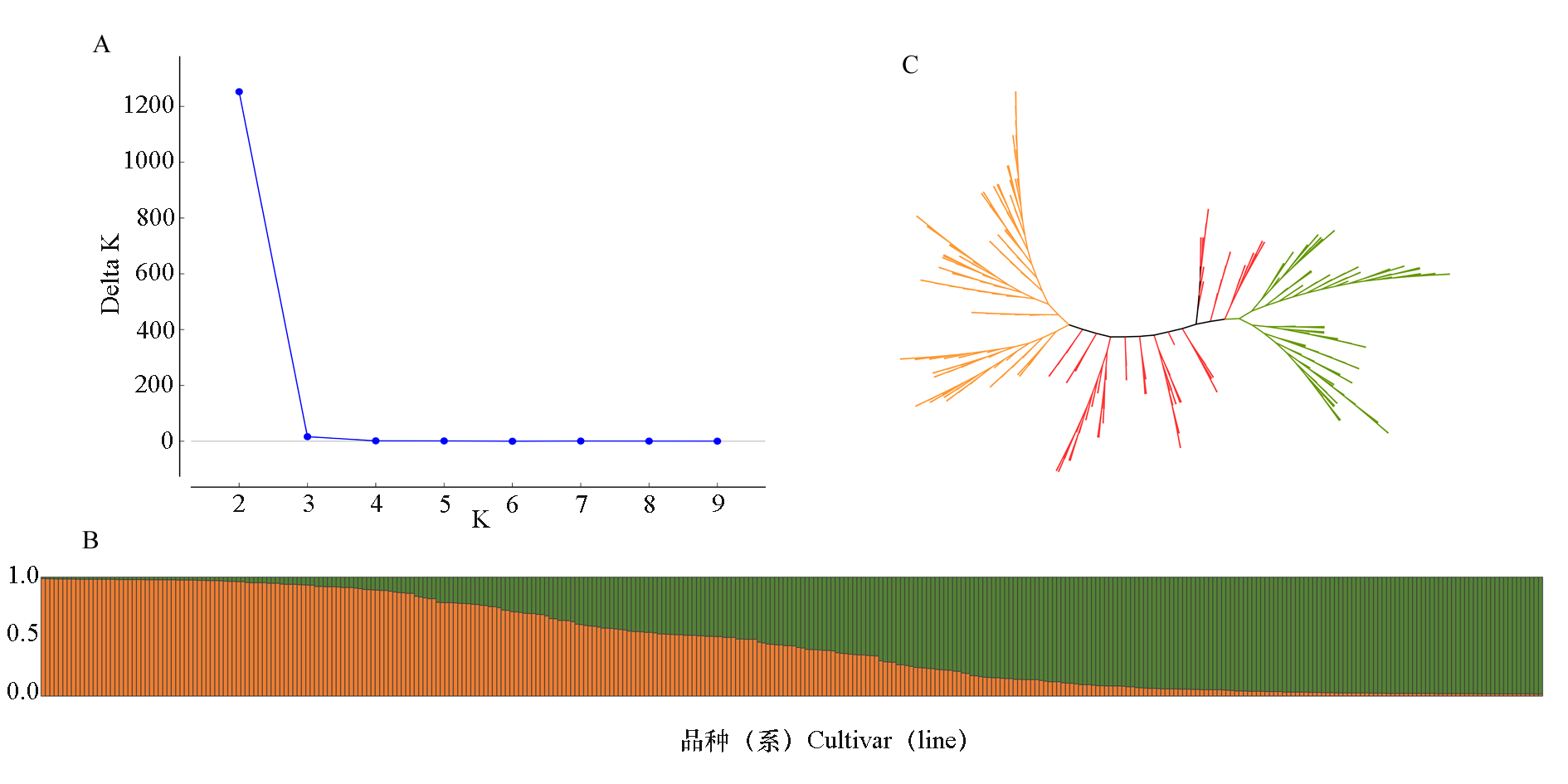
Fig. 3 Population structure and cluster tree of 342 Chinese chestnut cultivars(lines) A:Delta K value distribution of population structure analysis;B:Population structure at K = 2;C:UPGMA rootless tree of the tested Chinese chestnut cultivars(Orange represents group 1,green represents group 2,and red represents an intermediate types between group 1 and group 2)
| 群体 Pop | 样品数 Sample | 位点数 Na | 有效等位位点 Ne | Shannon’s 多样性指数 I | 观察杂合度 Ho | 期望杂合度 He |
|---|---|---|---|---|---|---|
| Pop1(华北品种群North China group) | 190 | 8.714 ± 0.772 | 3.675 ± 0.282 | 1.450 ± 0.083 | 0.577 ± 0.032 | 0.685 ± 0.030 |
| Pop2(西北品种群Northwest China group) | 10 | 4.857 ± 0.347 | 3.286 ± 0.210 | 1.289 ± 0.080 | 0.595 ± 0.044 | 0.658 ± 0.032 |
| Pop3(长江中下游品种群Middle and lower reaches of the Yangtze River group) | 100 | 7.810 ± 0.699 | 3.320 ± 0.294 | 1.347 ± 0.094 | 0.601 ± 0.042 | 0.642 ± 0.035 |
| Pop4(东南品种群 Southeast China group) | 18 | 6.286 ± 0.522 | 3.593 ± 0.339 | 1.391 ± 0.096 | 0.582 ± 0.050 | 0.662 ± 0.035 |
| Pop5(西南品种群 Southwest China group) | 24 | 6.381 ± 0.567 | 3.368 ± 0.309 | 1.462 ± 0.091 | 0.585 ± 0.043 | 0.703 ± 0.029 |
Table 3 The key genetic statistics of genetic diversity of five Chinese chestnut cultivar groups
| 群体 Pop | 样品数 Sample | 位点数 Na | 有效等位位点 Ne | Shannon’s 多样性指数 I | 观察杂合度 Ho | 期望杂合度 He |
|---|---|---|---|---|---|---|
| Pop1(华北品种群North China group) | 190 | 8.714 ± 0.772 | 3.675 ± 0.282 | 1.450 ± 0.083 | 0.577 ± 0.032 | 0.685 ± 0.030 |
| Pop2(西北品种群Northwest China group) | 10 | 4.857 ± 0.347 | 3.286 ± 0.210 | 1.289 ± 0.080 | 0.595 ± 0.044 | 0.658 ± 0.032 |
| Pop3(长江中下游品种群Middle and lower reaches of the Yangtze River group) | 100 | 7.810 ± 0.699 | 3.320 ± 0.294 | 1.347 ± 0.094 | 0.601 ± 0.042 | 0.642 ± 0.035 |
| Pop4(东南品种群 Southeast China group) | 18 | 6.286 ± 0.522 | 3.593 ± 0.339 | 1.391 ± 0.096 | 0.582 ± 0.050 | 0.662 ± 0.035 |
| Pop5(西南品种群 Southwest China group) | 24 | 6.381 ± 0.567 | 3.368 ± 0.309 | 1.462 ± 0.091 | 0.585 ± 0.043 | 0.703 ± 0.029 |

Fig. 4 The Fst values of pairwise populations during five Chestnut Cultivars groups Pop1 is the north China group;Pop2 is the northwest China group;Pop3 is the middle and lower reaches of the Yangtze River group;Pop4 is the southeast China group;Pop5 is the southwest China group.
| 变异来源 Source of variance | 自由度 df | 方差总和 SS | 平均方差 MS | 变异组分 Variance component | 变异百分率/% Variation | P |
|---|---|---|---|---|---|---|
| 种群间Among Pops | 4 | 134.102 | 33.526 | 0.249 | 3 | < 0.001 |
| 个体间Among Indiv | 337 | 2722.196 | 8.078 | 0.965 | 13 | < 0.001 |
| 个体内Within Indiv | 342 | 2102.500 | 6.148 | 6.148 | 84 | < 0.010 |
| 总量Total | 683 | 4958.798 | 7.362 | 100 |
Table 4 The AMOVA of five Chinese chestnut cultivar groups
| 变异来源 Source of variance | 自由度 df | 方差总和 SS | 平均方差 MS | 变异组分 Variance component | 变异百分率/% Variation | P |
|---|---|---|---|---|---|---|
| 种群间Among Pops | 4 | 134.102 | 33.526 | 0.249 | 3 | < 0.001 |
| 个体间Among Indiv | 337 | 2722.196 | 8.078 | 0.965 | 13 | < 0.001 |
| 个体内Within Indiv | 342 | 2102.500 | 6.148 | 6.148 | 84 | < 0.010 |
| 总量Total | 683 | 4958.798 | 7.362 | 100 |
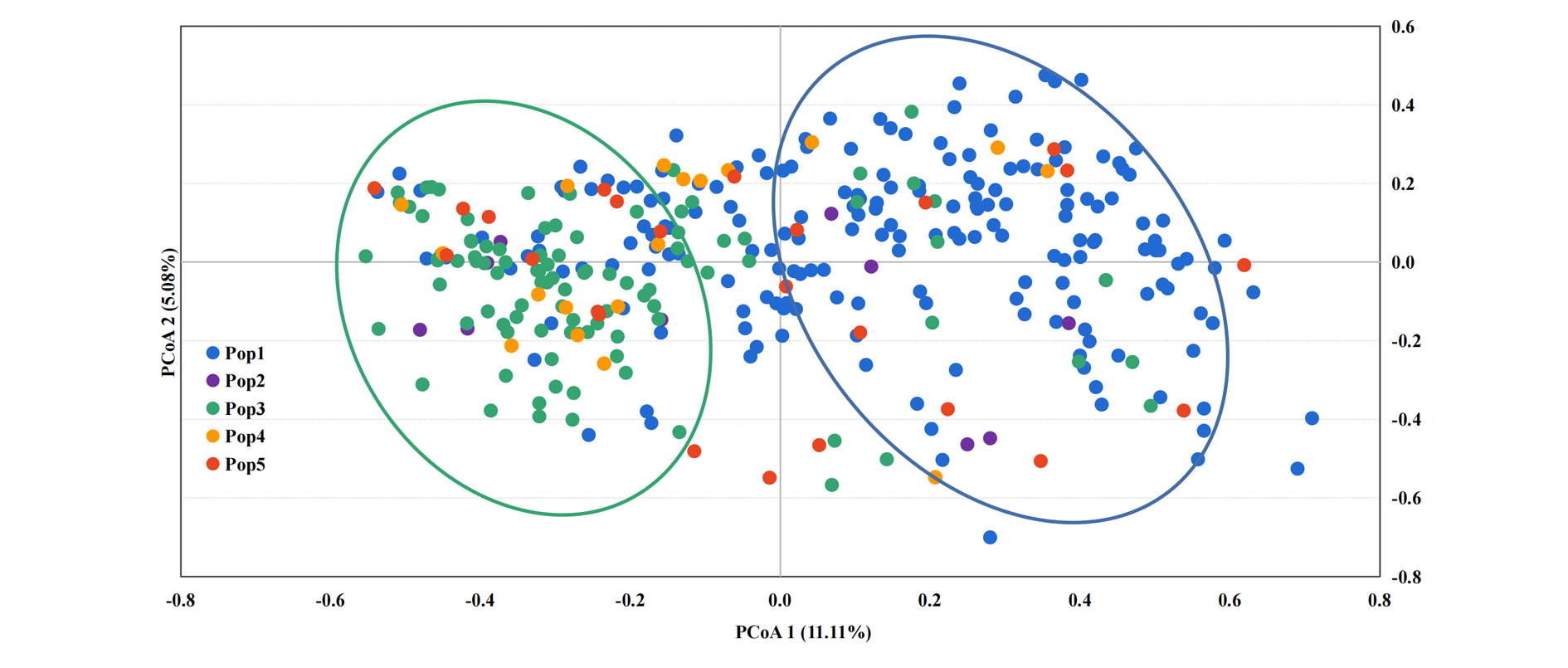
Fig. 5 Principal coordinate analysis of five Chinese chestnut cultivars groups Pop1 is the north China group;Pop2 is the northwest China group;Pop3 is the middle and lower reaches of the Yangtze River group;Pop4 is the southeast China group;Pop5 is the southwest China group.
| [1] |
Alcaraz M L, Hormaza J I. 2007. Molecular characterization and genetic diversity in an avocado collection of cultivars and local Spanish genotypes using SSRs. Hereditas, 144 (6):244-253.
doi: 10.1111/j.2007.0018-0661.02019x pmid: 18215247 |
| [2] | FAO. 2019. Value of agricultural production. http://faostat3.fao.org/. |
| [3] |
Falush D, Stephens M, Pritchard J K. 2003. Inference of population structure using multi-locus genotype data:Linked loci and correlated allele frequencies. Genetics, 164:1567-1587.
doi: 10.1093/genetics/164.4.1567 URL |
| [4] | Guo Jun, Zhu Jie, Xie Shangqian, Zhang Ye, Ye Beilei,Zheng Liyan,Ling Peng. 2020. Development of SSR molecular markers based on transcriptome and analysis of genetic relationship of germplasm resources in avocado. Acta Horticulturae Sinica, 47 (8):1552-1564. (in Chinese) |
| 郭俊, 朱婕, 谢尚潜, 张叶, 叶蓓蕾, 郑丽燕, 凌鹏. 2020. 油梨转录组SSR 分子标记开发与种质资源亲缘关系分析. 园艺学报, 47 (8):1552-1564. | |
| [5] | He Xu-dong, Zheng Ji-wei, Tian Xue-yao, Jiao Zhong-yi, Dou Quan-qin. 2021. Genetic relationship analysis and fingerprint construction of Carya illinoensis varieties. Forest Research, 34 (4):95-102. (in Chinese) |
| 何旭东, 郑纪伟, 田雪瑶, 教忠意, 窦全琴. 2021. 薄壳山核桃品种亲缘关系分析与指纹图谱构建. 林业科学研究, 34 (4):95-102. | |
| [6] |
Huang H W, Dane F, Norton J D. 1994. Allozyme diversity in Chinese Seguinii and American chestnut(Castanea spp.). Theoretical and Applied Genetics, 88 (8):981-985.
doi: 10.1007/BF00220805 pmid: 24186251 |
| [7] | Jiang Xibing, Tang Dan, Gong Bangchu, Lai Junsheng. 2015. Genetic diversity and association analysis of local cultivars of Chinese chestnut based on SSR markers. Acta Horticulturae Sinica, 42 (12):2478-2488. (in Chinese) |
| 江锡兵, 汤丹, 龚榜初, 赖俊声. 2015. 基于SSR标记的板栗地方品种遗传多样性与关联分析. 园艺学报, 42 (12):2478-2488. | |
| [8] | Jiang Xibing, Zhang Pingsheng, Xu Yang, Wu Conglian, Zhang Dongbei, Gong Bangchu, Wu Kaiyun, Lai Junsheng. 2021. Genetic diversity of F1 hybrids of chestnut based on SSR markers. Acta Horticulturae Sinica, 48 (5):897-907. (in Chinese) |
| 江锡兵, 章平生, 徐阳, 吴聪连, 张东北, 龚榜初, 吴开云, 赖俊声. 2021. 栗杂交F1代SSR标记遗传多样性分析. 园艺学报, 48 (5):897-907. | |
| [9] | Kubisiak T L, Nelson C D, Staton M E, Zhebentyayaeva T, Smith C, Olukolu B A, Fang C, Hebard F V, Anagnosakis S, Wheeler N, Sisco P H, Abbott A G, Sederoff R R. 2013. A transcriptome-based genetic map of Chinese chestnut(Castanea mollissima)and identification of regions of segmental homology with peach(Prunus persica). Tree Genetics & Genomes, 9:557-571. |
| [10] | Lang P, Dane F, Kubisiak T L. 2006. Phylogeny of Castanea(Fagaceae)based on chloroplasttrnT-L-F sequence data. Tree Genetics & Genomes, 2 (3):132-139. |
| [11] | Li Chao, Yang Ying, Chen Wei, Zheng Heyun, Liao Xinfu, Sun Yuping. 2022. Construction of DNA fingerprinting and clustering analysis with SSR markers for the muskmelon of Xizhoumi series. Acta Horticulturae Sinica, 49 (3):622-632. (in Chinese) |
| 李超, 杨英, 陈伟, 郑贺云, 廖新福, 孙玉萍. 2022. 西州密系列甜瓜SSR指纹图谱构建及聚类分析. 园艺学报, 49 (3):622-632. | |
| [12] | Liu Guo-bing, Cao Jun, Lan Yan-ping, Wang Jin-bao. 2017. Construction of SSR fingerprint on 33 ancient chestnut trees. Acta Agriculturae Universitatis Jiangxiensis, 39 (1):134-139. (in Chinese) |
| 刘国彬, 曹均, 兰彦平, 王金宝. 2017. 明清板栗古树指纹图谱的构建. 江西农业大学学报, 39 (1):134-139. | |
| [13] |
Liu K, Muse S V. 2005. PowerMarker:an integrated analysis environment for genetic marker analysis. Bioinformatics, 21:2128-2129.
doi: 10.1093/bioinformatics/bti282 URL |
| [14] |
Nie X H, Wang Z H, Liu N W, Song Li, Yan B Q, Xing Y, Zhang Q, Fang K F, Zhao Y L, Chen X, Wang G P, Qin L, Cao Q Q. 2021. Fingerprinting 146 Chinese chestnut accessions and selecting a core collection using SSR markers. Journal of Integrative Agriculture, 20 (5):1277-1286.
doi: 10.1016/S2095-3119(20)63400-1 |
| [15] | Nie Xing-hua, Zheng Rui-jie, Zhao Yong-lian, Cao Qing-qin, Qin Ling, Xing Yu. 2021. Genetic diversity evaluation of Castanea in China based on fluorescently labeled SSR. Scientia Agricultura Sinica, 54 (8):1739-1750. (in Chinese) |
| 聂兴华, 郑瑞杰, 赵永廉, 曹庆芹, 秦岭, 邢宇. 2021. 利用荧光SSR分子标记评估中国栗属植物遗传多样性. 中国农业科学, 54 (8):1739-1750. | |
| [16] |
Peakall R, Smouse P E. 2006. GENALEX 6:genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes, 6:288-295.
doi: 10.1111/j.1471-8286.2005.01155.x URL |
| [17] | Riva G A, Cyril J C, Yuval E, Leora S. 2000. Simple Sequence Repeats in Escherichia coli:abundance,distribution,composition,and polymorphism. Genome Research, 10 (1):62-71. |
| [18] | Rod P, Smouse P E. 2012. GenAlEx 6.5:genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics,(19):2537-2539. |
| [19] |
Selkoe K A, Toonen R J. 2006. Microsatellites for ecologists:a practical guide to using and evaluating microsatellite markers. Ecology Letters, 9:615-629.
doi: 10.1111/j.1461-0248.2006.00889.x URL |
| [20] |
Taberlet P, Luikart G. 1999. Non-invasive genetic sampling and individual identification. Biological Journal of the Linnean Society, 68:41-55.
doi: 10.1111/j.1095-8312.1999.tb01157.x URL |
| [21] |
Tian Hua, Kang Ming, Yao Xiao-Hong,Huang Hong-Wen. 2009. Genetic diversity in natural populations of Castanea mollissima inferred from nuclear SSR markers. Biodiversity Science, 17 (3):296-302. (in Chinese)
doi: 10.3724/SP.J.1003.2009.09043 URL |
|
田华, 康明, 李丽, 姚小洪, 黄宏文. 2009. 中国板栗自然居群微卫星(SSR)遗传多样性. 生物多样性, 17 (3):296-302.
doi: 10.3724/SP.J.1003.2009.09043 |
|
| [22] | UPOV. 2007. Guidelines for DNA-profiling:Molecular marker selection and database construction/BMT Guidelines(proj.9). Geneva,Switzerland:3-4. |
| [23] |
Waits L P, Luikart G, Taberlet P. 2001. Estimating the probability of identity among genotypes in natural populations:cautions and guidelines. Molecular Ecology, 10:249-256.
doi: 10.1046/j.1365-294x.2001.01185.x pmid: 11251803 |
| [24] |
Wright S. 1931. Evolution in Mendelian populations. Genetics, 16:97-159.
doi: 10.1093/genetics/16.2.97 pmid: 17246615 |
| [25] | Ye Shu-tao, Fang Zhou, Li Ying-lin, Wu Jun-jian, Zheng Guo-hua, Chen Hui, Li Yu. 2021. The SSR variation for cultivated population of Castanea henryi variety ‘Youzhen’. Molecular Plant Breeding, 6:1-9. (in Chinese) |
| 叶树涛, 方周, 李颖林, 吴钧剑, 郑国华, 陈辉, 李煜. 2021. 锥栗农家品种‘油榛’栽培群体的SSR变异分析. 分子植物育种, 6:1-9. | |
| [26] | Zhang He-yu, Liang Wei-jian. 2005. Chinese fruit tree,Chinese chestnut,hazelnut rolls. Beijing: China Forestry Publishing House. (in Chinese) |
| 张宇和, 柳鎏, 梁维坚. 2005. 中国果树志. 板栗,榛子卷. 北京: 中国林业出版社. | |
| [27] | Zhang Xin-fang, Zhang Shu-hang, Li Ying, Wang Guang-peng. 2020. Genetic diversity analysis of chestnut germplasm in Yanshan region based on SSR markers. Journal of China Agricultural University, 25 (4):61-71. (in Chinese) |
| 张馨方, 张树航, 李颖, 王广鹏. 2020. 基于SSR标记的燕山板栗种质资源遗传多样性分析. 中国农业大学学报, 25 (4):61-71. | |
| [28] | Zhou Lian-di. 2005. Study on genetic diversity of germplasm resources in Castanea mollissima[Ph. D. Dissertation]. China Agricultural University. (in Chinese) |
| 周连第. 2005. 板栗种质资源遗传多样性研究[博士论文]. 北京: 中国农业大学. |
| [1] | WANG Rui, HONG Wenjuan, LUO Hua, ZHAO Lina, CHEN Ying, and WANG Jun, . Construction of SSR Fingerprints of Pomegranate Cultivars and Male Parent Identification of Hybrids [J]. Acta Horticulturae Sinica, 2023, 50(2): 265-278. |
| [2] | WANG Mengmeng, SUN Deling, CHEN Rui, YANG Yingxia, ZHANG Guan, LÜ Mingjie, WANG Qian, XIE Tianyu, NIU Guobao, SHAN Xiaozheng, TAN Jin, and YAO Xingwei, . Construction and Evaluation of Cauliflower Core Collection [J]. Acta Horticulturae Sinica, 2023, 50(2): 421-431. |
| [3] | LIU Yiping, NI Menghui, WU Fangfang, LIU Hongli, HE Dan, KONG Dezheng. Association Analysis of Organ Traits with SSR Markers in Lotus(Nelumbo nucifera) [J]. Acta Horticulturae Sinica, 2023, 50(1): 103-115. |
| [4] | LI Wenting, LI Cuixiao, LIN Xiaoqing, ZHENG Yongqin, ZHENG Zheng, DENG Xiaoling. Population Genetic Structure of Xanthomonas citri pv. citri in Guangdong Province Based on the STR Locus [J]. Acta Horticulturae Sinica, 2022, 49(6): 1233-1246. |
| [5] | ZHANG Meng, SHAN Yuying, YANG Yebo, ZHAI Feifei, WANG Zhaoshan, JU Guansheng, SUN Zhenyuan, LI Zhenjian. AFLP Analysis of Genetic Resources of Dendrobium from China [J]. Acta Horticulturae Sinica, 2022, 49(6): 1339-1350. |
| [6] | YANG Yi, LI Tingyao, LI Guojing, CHEN Hancai, SHEN Zhuo, ZHOU Xuan, WU Zengxiang, WU Xinyi, ZHANG Yan. Development and Application of Insertion-Deletion(InDel)Markers in Asparagus Bean Based on Whole Genome Re-sequencing Data [J]. Acta Horticulturae Sinica, 2022, 49(4): 778-790. |
| [7] | LI Chao, YANG Ying, CHEN Wei, ZHENG Heyun, LIAO Xinfu, SUN Yuping. Construction of DNA Fingerprinting and Clustering Analysis with SSR Markers for the Muskmelon of Xizhoumi Series [J]. Acta Horticulturae Sinica, 2022, 49(3): 622-632. |
| [8] | LI Shifan, SHAO Yuchun, QI Haonan, YANG Lan, LI Xiaowei, XU Bingliang, CHEN Yahan. Detection and Genetic Diversity Analysis of Apple Necrotic Mosaic Virus in Gansu Province [J]. Acta Horticulturae Sinica, 2022, 49(11): 2431-2438. |
| [9] | CHEN Mingkun, CHEN Lu, SUN Weihong, MA Shanhu, LAN Siren, PENG Donghui, LIU Zhongjian, AI Ye. Genetic Diversity Analysis and Core Collection of Cymbidium ensifolium Germplasm Resources [J]. Acta Horticulturae Sinica, 2022, 49(1): 175-186. |
| [10] | SONG Yun, JIA Mengjun, CAO Yaping, LI Zheng, HE Jiaxin, WANG Yongfei, ZHANG Xinrui, QIAO Yonggang. Analysis on Chloroplast Genomic Characteristics of Forsythia suspensa [J]. Acta Horticulturae Sinica, 2022, 49(1): 187-199. |
| [11] | LI Yanhong, NIE Jun, ZHENG Jinrong, TAN Delong, ZHANG Changyuan, SHI Liangliang, XIE Yuming. Genetic Diversity Analysis and Multivariate Evaluation of Cherry Tomato by Phenotypic Traits in South China [J]. Acta Horticulturae Sinica, 2021, 48(9): 1717-1730. |
| [12] | WANG Xin, LI Mingyang, TIAN Lin, LIU Dongyun. ISSR and rDNA-ITS Sequence Analysis of the Genetic Relationship of Clematis in Hebei Province [J]. Acta Horticulturae Sinica, 2021, 48(9): 1755-1767. |
| [13] | ZHAO Yujing, LU Yin, FENG Daling, LUO Lei, WU Fang, YANG Rui, CHEN Yueqi, SUN Xiaoxue, WANG Yanhua, CHEN Xueping, SHEN Shuxing, LUO Shuangxia, ZHAO Jianjun. Association Analysis Between Background Selection InDel Markers and Important Agronomic Traits in Inbred Lines of Chinese Cabbage [J]. Acta Horticulturae Sinica, 2021, 48(7): 1282-1294. |
| [14] | DING Yunhua, Budahn Holger, ZHAO Hong, ZHAO Xiuyun. Accurate Identification of Radish Chromosome F in the Backcross Progeny of Brassica rapa and Rape-radish Chromosome F Addition Line [J]. Acta Horticulturae Sinica, 2021, 48(7): 1295-1303. |
| [15] | ZHAO Qing, DU Zhenzhen, LI Xixiang, SONG Jiangping, ZHANG Xiaohui, YANG Wenlong, JIA Huixia, WANG Haiping. Genetic Diversity of Garlic Germplasm Resources Based on SSRseq Molecular Markers [J]. Acta Horticulturae Sinica, 2021, 48(7): 1397-1408. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd