
Acta Horticulturae Sinica ›› 2022, Vol. 49 ›› Issue (8): 1815-1832.doi: 10.16420/j.issn.0513-353x.2021-0243
• Review • Previous Articles Next Articles
WANG Yongjian1, KONG Junhua1, FAN Peige1, LIANG Zhenchang1, JIN Xiuliang2, LIU Buchun3, DAI Zhanwu1,*( )
)
Received:2021-06-10
Revised:2022-01-29
Online:2022-08-25
Published:2022-09-05
Contact:
DAI Zhanwu
E-mail:zhanwu.dai@ibcas.ac.cn
CLC Number:
WANG Yongjian, KONG Junhua, FAN Peige, LIANG Zhenchang, JIN Xiuliang, LIU Buchun, DAI Zhanwu. Grape Phenome High-throughput Acquisition and Analysis Methods:A Review[J]. Acta Horticulturae Sinica, 2022, 49(8): 1815-1832.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2021-0243
| 传感器类型 Sensor type | 尺度 Scale | 表型指标 Phenotype trait | 数据获取环境 Data acquisition environment | 优势 Advantage | 劣势 Disadvantage | 参考文献 Reference |
|---|---|---|---|---|---|---|
| RGB相机 RGB camera | 冠层,植株,器官 Canopy,plant,organ | 投影面积,器官数量,形态结构,颜色纹理 Project area,organ quantity,morphological structure,color texture | 可控环境,田间 Controlled environment,field | 成本低;分辨率高;集成方便;适用于不同尺度 Cost-effective;high-resolution;easy integration;suitable for multi-scales | 只能获取可见光波段;图像质量受环境影响;仅能获取相对值;二维图像 Only visible wavelengths can be captured;environmental impact on image quality;only get the relative value;2D image | Aquino et al., |
| 光谱相机 Spectral Camera | 冠层,植株,器官 Canopy,plant,organ | 投影面积,器官识别,形态结构,颜色,纹理 Project area,organ identification,morphological structure,color,texture | 可控环境,田间 Controlled environment,field | 可获取非可见光波段图像;集成方便;适用于不同尺度 Obtain images in non-visible band;easy integration;suitable for multi-scales | 成本较高;图像分辨率低于RGB相机;受环境影响;获取过程需要多次校正 High cost;image resolution is lower than RGB camera;affected by the environment;the acquisition process requires frequently calibrated | Rodríguez-Pulido et al., |
| 深度相机 Depth camera | 冠层 Canopy | 植株分割,器官识别 Plant segmentation,organ identify | 可控环境,田间 Controlled environment,field | 三维成像;成本可控;集成方便 3D image;cost effective;easy integration | 分辨率较低;受环境影响;仅能获取相对值 Low resolution;affected by the environments;only get the relative value | Lopes et al., |
| 三维扫描仪 3D scanner | 冠层,器官 Canopy,organ | 三维结构,空间分布 3D architecture,spatial distribution | 可控环境,田间 Controlled environment,field | 三维点云信息;绝对距离;精度高;受环境影响低 3D point clouds;Absolute distance;High-precision;environment-stable | 成本较高;原始数据体量较大;数据获取及处理耗时长 High cost;large raw data;time consuming for data acquisition and processing | Schöler & Steinhage, |
| X射线断层扫描 X-ray tomography | 器官 Organ | 内部密度差异结构,三维结构 Internal density differential structure,3D architecture | 可控环境 Controlled environment | 三维内部结构信息;绝对距离;精度高 Internal 3D information;absolute distance;high precision | 成本极高;原始数据体量大;数据获取及处理耗时长;样品预处理复杂 Very high cost;large raw data;time consuming for data acquisition and processing;complicated sample pretreatment. | Li et al., |
Table 1 Comparison of different phenotypic acquisition sensors
| 传感器类型 Sensor type | 尺度 Scale | 表型指标 Phenotype trait | 数据获取环境 Data acquisition environment | 优势 Advantage | 劣势 Disadvantage | 参考文献 Reference |
|---|---|---|---|---|---|---|
| RGB相机 RGB camera | 冠层,植株,器官 Canopy,plant,organ | 投影面积,器官数量,形态结构,颜色纹理 Project area,organ quantity,morphological structure,color texture | 可控环境,田间 Controlled environment,field | 成本低;分辨率高;集成方便;适用于不同尺度 Cost-effective;high-resolution;easy integration;suitable for multi-scales | 只能获取可见光波段;图像质量受环境影响;仅能获取相对值;二维图像 Only visible wavelengths can be captured;environmental impact on image quality;only get the relative value;2D image | Aquino et al., |
| 光谱相机 Spectral Camera | 冠层,植株,器官 Canopy,plant,organ | 投影面积,器官识别,形态结构,颜色,纹理 Project area,organ identification,morphological structure,color,texture | 可控环境,田间 Controlled environment,field | 可获取非可见光波段图像;集成方便;适用于不同尺度 Obtain images in non-visible band;easy integration;suitable for multi-scales | 成本较高;图像分辨率低于RGB相机;受环境影响;获取过程需要多次校正 High cost;image resolution is lower than RGB camera;affected by the environment;the acquisition process requires frequently calibrated | Rodríguez-Pulido et al., |
| 深度相机 Depth camera | 冠层 Canopy | 植株分割,器官识别 Plant segmentation,organ identify | 可控环境,田间 Controlled environment,field | 三维成像;成本可控;集成方便 3D image;cost effective;easy integration | 分辨率较低;受环境影响;仅能获取相对值 Low resolution;affected by the environments;only get the relative value | Lopes et al., |
| 三维扫描仪 3D scanner | 冠层,器官 Canopy,organ | 三维结构,空间分布 3D architecture,spatial distribution | 可控环境,田间 Controlled environment,field | 三维点云信息;绝对距离;精度高;受环境影响低 3D point clouds;Absolute distance;High-precision;environment-stable | 成本较高;原始数据体量较大;数据获取及处理耗时长 High cost;large raw data;time consuming for data acquisition and processing | Schöler & Steinhage, |
| X射线断层扫描 X-ray tomography | 器官 Organ | 内部密度差异结构,三维结构 Internal density differential structure,3D architecture | 可控环境 Controlled environment | 三维内部结构信息;绝对距离;精度高 Internal 3D information;absolute distance;high precision | 成本极高;原始数据体量大;数据获取及处理耗时长;样品预处理复杂 Very high cost;large raw data;time consuming for data acquisition and processing;complicated sample pretreatment. | Li et al., |
| 表型提取方法 Phenotyping extraction method | 原始数据类型 Raw data type | 表型指标 Phenotyping trait | 分析平台 Analysis platform | 参考文献 Reference |
|---|---|---|---|---|
| 图像分割 Image segmentation | RGB图像 RGB Image | LAI、果穗数、芽数、修剪量、孔隙度、冠层投影面积、果穗紧实度、果粒数、颜色纹理、花粉数 LAI,cluster quantity,bud quantity,pruning weight,porosity,canopy projection area,cluster compactness,berry quantity,color texture,pollen quantity | Matlab;ImageJ;OpenCV | Aquino et al., |
| 机器学习 Machine learning | RGB图像 RGB image | 器官识别、叶片病害识别 Organ identification,leaf disease identification | AlexNet;MobileNets; ShuffleNet V1;ShuffleNet V2 | Botterill et al., |
| 光谱分析 Spectrum analysis | 光谱图像 Spectral image | 叶片氮含量、病虫害、种子质量 Leaf nitrogen content,pests and diseases,seed quality | Matlab;ImageJ;OpenCV… | Rodríguez-Pulido et al., di Gennaro et al., |
| 三维重建 3D reconstruction | RGB图像 RGB image | 冠层体积、果穗体积、果穗三维形态、分支结构 Canopy volume,cluster volume,cluster 3D architecture,branch architecture | Agisoft Photoscan; structure-from-motion (SFM) | Kicherer et al., |
| 点云处理 Point cloud processing | 点云数据 Point cloud data | 果穗三维空间结构、果粒数、果穗紧实度 3D cluster spatial architecture,berry quantity,cluster compactness | CloudCompare;Artec Studio;Geomagic Studio;... | Schöler & Steinhage, |
Table 2 Comparison of different phenotypic analysis methods
| 表型提取方法 Phenotyping extraction method | 原始数据类型 Raw data type | 表型指标 Phenotyping trait | 分析平台 Analysis platform | 参考文献 Reference |
|---|---|---|---|---|
| 图像分割 Image segmentation | RGB图像 RGB Image | LAI、果穗数、芽数、修剪量、孔隙度、冠层投影面积、果穗紧实度、果粒数、颜色纹理、花粉数 LAI,cluster quantity,bud quantity,pruning weight,porosity,canopy projection area,cluster compactness,berry quantity,color texture,pollen quantity | Matlab;ImageJ;OpenCV | Aquino et al., |
| 机器学习 Machine learning | RGB图像 RGB image | 器官识别、叶片病害识别 Organ identification,leaf disease identification | AlexNet;MobileNets; ShuffleNet V1;ShuffleNet V2 | Botterill et al., |
| 光谱分析 Spectrum analysis | 光谱图像 Spectral image | 叶片氮含量、病虫害、种子质量 Leaf nitrogen content,pests and diseases,seed quality | Matlab;ImageJ;OpenCV… | Rodríguez-Pulido et al., di Gennaro et al., |
| 三维重建 3D reconstruction | RGB图像 RGB image | 冠层体积、果穗体积、果穗三维形态、分支结构 Canopy volume,cluster volume,cluster 3D architecture,branch architecture | Agisoft Photoscan; structure-from-motion (SFM) | Kicherer et al., |
| 点云处理 Point cloud processing | 点云数据 Point cloud data | 果穗三维空间结构、果粒数、果穗紧实度 3D cluster spatial architecture,berry quantity,cluster compactness | CloudCompare;Artec Studio;Geomagic Studio;... | Schöler & Steinhage, |
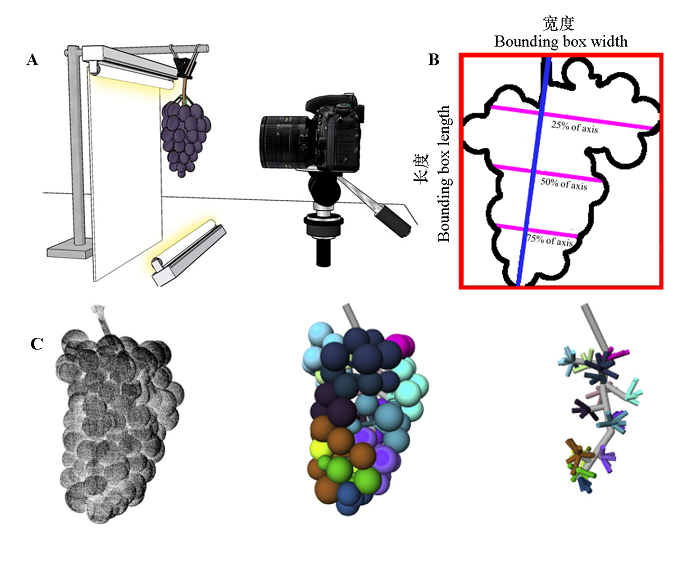
Fig. 4 Methods for obtaining grape cluster phenotype A:Schematic diagram of grape cluster image acquisition facility(Underhill et al.,2020b);B:Diagram of grape cluster morphological parameters (Underhill et al.,2020b);C:3D reconstruction and phenotypic analysis of grape cluster. The left image is the input point cloud acquired by laser scanning,the middle image is the reconstruction of berries,and the right image is the branch structure of grape cluster(Schöler & Steinhage,2015).
| [1] |
Aquino A, Barrio I, Diago M P, Millan B, Tardaguila J. 2018a. Vitisberry:an android-smartphone application to early evaluate the number of grapevine berries by means of image analysis. Computers and Electronics in Agriculture, 148:19-28.
doi: 10.1016/j.compag.2018.02.021 URL |
| [2] |
Aquino A, Millan B, Diago M P, Tardaguila J. 2018b. Automated early yield prediction in vineyards from on-the-go image acquisition. Computers and Electronics in Agriculture, 144:26-36.
doi: 10.1016/j.compag.2017.11.026 URL |
| [3] | Aquino A, Millan B, Gaston D, Diago M P, Tardaguila J. 2015. Vitisflower(r):development and testing of a novel android-smartphone application for assessing the number of grapevine flowers per inflorescence using artificial vision techniques. Sensors(Basel), 15:21204-21218. |
| [4] |
Badouin H, Velt A, Gindraud F, Flutre T, Dumas V, Vautrin S, Marande W, Corbi J, Sallet E, Ganofsky J. 2020. The wild grape genome sequence provides insights into the transition from dioecy to hermaphroditism during grape domestication. Genome Biology, 21:1-24.
doi: 10.1186/s13059-019-1906-x URL |
| [5] |
Baumgartner A, Donahoo M, Chitwood D H, Peppe D J. 2020. The influences of environmental change and development on leaf shape in vitis. American Journal of Botany, 107:676-688.
doi: 10.1002/ajb2.1460 pmid: 32270876 |
| [6] |
Bendel N, Kicherer A, Backhaus A, Kluck H C, Seiffert U, Fischer M, Voegele R T, Topfer R. 2020. Evaluating the suitability of hyper- and multispectral imaging to detect foliar symptoms of the grapevine trunk disease esca in vineyards. Plant Methods, 16:142.
doi: 10.1186/s13007-020-00685-3 pmid: 33101451 |
| [7] |
Bernardo R. 2020. Reinventing quantitative genetics for plant breeding:something old,something new,something borrowed,something blue. Heredity, 125:375-385.
doi: 10.1038/s41437-020-0312-1 URL |
| [8] |
Botterill T, Paulin S, Green R, Williams S, Lin J, Saxton V, Mills S, Chen X, Corbett-Davies S. 2017. A robot system for pruning grape vines. Journal of Field Robotics, 34:1100-1122.
doi: 10.1002/rob.21680 URL |
| [9] | Bryson A E, Wilson B M, Mullins J, Dong W, Bahmani K, Bornowski N, Chiu C, Engelgau P, Gettings B, Gomezcano F. 2020. Composite modeling of leaf shape along shoots discriminates vitis species better than individual leaves. Applications in Plant Sciences, 8 (12):e11404. |
| [10] |
Chen J, Wang N, Fang L C, Liang Z C, Li S H, Wu B H. 2015. Construction of a high-density genetic map and QTLs mapping for sugars and acids in grape berries. BMC Plant Biology, 15:1-14.
doi: 10.1186/s12870-014-0410-4 URL |
| [11] | Cheng Chao-hua, Tang Qing, Deng Can-hui, Dai Zhi-gang, Xu Ying, Yang Ze-mao, Liu Chan, Wu Shan, Su Jiang-guang. 2020. Application of phenomics and multiomics joint analysis in accurate identification of plant germplasm resources. Molecular Plant Breeding, 18:2747-2753. (in Chinese) |
| 程超华, 唐蜻, 邓灿辉, 戴志刚, 许英, 杨泽茂, 刘婵, 吴姗, 粟建光. 2020. 表型组学及多组学联合分析在植物种质资源精准鉴定中的应用. 分子植物育种, 18:2747-2753. | |
| [12] |
Chitwood D H. 2020. The shapes of wine and table grape leaves:an ampelometric study inspired by the methods of pierre galet. Plants,People,Planet, 3:155-170.
doi: 10.1002/ppp3.10157 URL |
| [13] |
Chitwood D H, Klein L L, O'Hanlon R, Chacko S, Greg M, Kitchen C, Miller A J, Londo J P. 2016a. Latent developmental and evolutionary shapes embedded within the grapevine leaf. New Phytologist, 210:343-355.
doi: 10.1111/nph.13754 URL |
| [14] |
Chitwood D H, Ranjan A, Martinez C C, Headland L R, Thiem T, Kumar R, Covington M F, Hatcher T, Naylor D T, Zimmerman S, Downs N, Raymundo N, Buckler E S, Maloof J N, Aradhya M, Prins B, Li L, Myles S, Sinha N R. 2014. A modern ampelography:a genetic basis for leaf shape and venation patterning in grape. Plant Physiology, 164:259-272.
doi: 10.1104/pp.113.229708 pmid: 24285849 |
| [15] |
Chitwood D H, Rundell S M, Li D Y, Woodford Q L, Tommy T Y, Lopez J R, Greenblatt D, Kang J, Londo J P. 2016b. Climate and developmental plasticity:interannual variability in grapevine leaf morphology. Plant Physiology, 170:1480-1491.
doi: 10.1104/pp.15.01825 URL |
| [16] |
Comar A, Burger P, de Solan B, Baret F D R, Daumard F, Hanocq J O. 2012. A semi-automatic system for high throughput phenotyping wheat cultivars in-field conditions:description and first results. Funct Plant Biol, 39:914-924.
doi: 10.1071/FP12065 URL |
| [17] |
CooMbe B G, McCarthy M. 2000. Dynamics of grape berry growth and physiology of ripening. Australian Journal of Grape Wine Research, 6:131-135.
doi: 10.1111/j.1755-0238.2000.tb00171.x URL |
| [18] |
Coviello L, Cristoforetti M, Jurman G, Furlanello C. 2020. Gbcnet:in-field grape berries counting for yield estimation by dilated CNNs. Applied Sciences, 10 (14):4870.
doi: 10.3390/app10144870 URL |
| [19] |
Cubero S, Diago M P, Blasco J, Tardáguila J, Prats-Montalbán J M, Ibáñez J, Tello J, Aleixos N. 2015. A new method for assessment of bunch compactness using automated image analysis. Australian Journal of Grape Wine Research, 21:101-109.
doi: 10.1111/ajgw.12118 URL |
| [20] | Dalal A, Shenhar I, Bourstein R, Mayo A, Grunwald Y, Averbuch N, Attia Z, Wallach R, Moshelion M. 2020. A telemetric,gravimetric platform for real-time physiological phenotyping of plant-environment interactions. Journal of Visualized Experiments, 162:e61280. |
| [21] |
di Gennaro S F, Toscano P, Cinat P, Berton A, Matese A. 2019. A low-cost and unsupervised image recognition methodology for yield estimation in a vineyard. Front Plant Sci, 10:559.
doi: 10.3389/fpls.2019.00559 URL |
| [22] |
Divilov K, Barba P, Cadle-Davidson L, Reisch B I. 2018. Single and multiple phenotype qtl analyses of downy mildew resistance in interspecific grapevines. Theoretical and Applied Genetics, 131:1133-1143.
doi: 10.1007/s00122-018-3065-y URL |
| [23] |
Doligez A, Bertrand Y, Farnos M, Grolier M, Romieu C, Esnault F, Dias S, Berger G, François P, Pons T. 2013. New stable qtls for berry weight do not colocalize with qtls for seed traits in cultivated grapevine(Vitis vinifera L.). BMC Plant Biology, 13:1-16.
doi: 10.1186/1471-2229-13-1 URL |
| [24] | Dong J, Burnham J G, Boots B, Rains G, Dellaert F. 2017. 4D crop monitoring:Spatio-temporal reconstruction for agriculture//2017 IEEE International Conference on Robotics and Automation(ICRA). IEEE,3878-3885. |
| [25] | Dong J, Carlone L, Rains G C, Coolong T, Dellaert F. 2014. 4D mapping of fields using autonomous ground and aerial vehicles//ASABE Annual International Meeting. American Society of Agricultural and Biological Engineers:1-7. |
| [26] | Fang Jing-gui, Liu Chong-huai. 2014. Molecular biology of grape. Beijing: Science Press:46-47. (in Chinese) |
| 房经贵, 刘崇怀. 2014. 葡萄分子生物学. 北京: 科学出版社:46-47. | |
| [27] | FAO, 2009. https://www.fao.org/faostat/en/#data/QCL. |
| [28] |
Feldmann M J, Hardigan M A, Famula R A, Lopez C M, Tabb A, Cole G S, Knapp S J. 2020. Multi-dimensional machine learning approaches for fruit shape phenotyping in strawberry. GigaScience, 9 (5):giaa030.
doi: 10.1093/gigascience/giaa030 URL |
| [29] |
Fernandez L, Torregrosa L, Segura V, Bouquet A, Martinez-Zapater J M. 2010. Transposon-induced gene activation as a mechanism generating cluster shape somatic variation in grapevine. The Plant Journal, 61:545-557.
doi: 10.1111/j.1365-313X.2009.04090.x pmid: 19947977 |
| [30] |
Fournier-Level A, Lacombe T, Le Cunff L, Boursiquot J-M, This P. 2010. Evolution of the VvMybA gene family,the major determinant of berry colour in cultivated grapevine(Vitis vinifera L.). Heredity, 104:351-362.
doi: 10.1038/hdy.2009.148 pmid: 19920856 |
| [31] | Gao M, Lu T F. 2006. Image processing and analysis for autonomous grapevine pruning//International Conference on Mechatronics and Automation. 922-927. |
| [32] | Ge Meng-qing, Dong Tian-yu, Fan Xiu-cai, Liu Chong-huai. 2020a. Investigation on the weight,volume and related characters of grape berry. Sino-Overseas Grapevine & Wine,(1):22-30. (in Chinese) |
| 葛孟清, 董天宇, 樊秀彩, 刘崇怀. 2020a. 葡萄浆果质量、体积及相关性状的调查. 中外葡萄与葡萄酒,(1):22-30. | |
| [33] | Ge Meng-qing, Dong Tian-yu, Zheng Ting, Shangguan Ling-fei, Fang Jing-gui, Liu Chong-huai. 2020b. Investigation and difference analysis on characters of fruit size of 48 grape(Vitis spp.) cultivars. Journal of Plant Resources and Environment, 29:19-27. (in Chinese) |
| 葛孟清, 董天宇, 郑婷, 上官凌飞, 房经贵, 刘崇怀. 2020b. 48个葡萄品种果实大小粒性状调查及差异分析. 植物资源与环境学报, 29:19-27. | |
| [34] | Guo Qing-hua, Yang Wei-cai, Wu Fang-fang, Pang Shu-xin, Jin Shi-chao, Chen Fan, Wang Xiu-jie. 2018. High-throughput crop phenotyping:accelerators for development of breeding and precision agriculture. Bulletin of Chinese Academy of Sciences, 33:940-946. (in Chinese) |
| 郭庆华, 杨维才, 吴芳芳, 庞树鑫, 金时超, 陈凡, 王秀杰. 2018. 高通量作物表型监测:育种和精准农业发展的加速器. 中国科学院院刊, 33:940-946. | |
| [35] |
Hacking C, Poona N, Poblete-Echeverría C. 2020. Vineyard yield estimation using 2-D proximal sensing:a multitemporal approach. OENO One, 54:793-812.
doi: 10.20870/oeno-one.2020.54.4.3361 URL |
| [36] | Hammer G, Messina C, van Oosterom E, Chapman S, Singh V, Borrell A, Jordan D, Cooper M. 2016. Molecular breeding for complex adaptive traits:how integrating crop ecophysiology and modelling can enhance efficiency. Crop Systems Biology,Springer,Cham:147-162. |
| [37] |
Hämmerle M, Höfle B. 2014. Effects of reduced terrestrial lidar point density on high-resolution grain crop surface models in precision agriculture. Sensors, 14:24212-24230.
doi: 10.3390/s141224212 pmid: 25521383 |
| [38] |
Hed B, Ngugi H K, Travis J W. 2009. Relationship between cluster compactness and bunch rot in vignoles grapes. Plant Disease, 93:1195-1201.
doi: 10.1094/PDIS-93-11-1195 URL |
| [39] | Herzog K, Roscher R, Wieland M, Kicherer A, Labe T, Forstner W, Kuhlmann H, Topfer R. 2014. Initial steps for high-throughput phenotyping in vineyards. Vitis, 53:1-8. |
| [40] |
Herzog K, Wind R, Topfer R. 2015. Impedance of the grape berry cuticle as a novel phenotypic trait to estimate resistance to Botrytis cinerea. Sensors, 15:12498-12512.
doi: 10.3390/s150612498 pmid: 26024417 |
| [41] |
Huther P, Schandry N, Jandrasits K, Bezrukov I, Becker C. 2020. Rsis,an automated workflow for top-view plant phenomics using semantic segmentation of leaf states. Plant Cell, 32 (2):3674-3688.
doi: 10.1105/tpc.20.00318 URL |
| [42] |
Ivorra E, Sánchez A J, Camarasa J G, Diago M P, Tardaguila J. 2015. Assessment of grape cluster yield components based on 3D descriptors using stereo vision. Food Control, 50:273-282.
doi: 10.1016/j.foodcont.2014.09.004 URL |
| [43] |
Jaillon O, Aury J-M, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C. 2007. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature, 449:463.
doi: 10.1038/nature06148 URL |
| [44] |
Jin X, Zarco-Tejada P, Schmidhalter U, Reynolds M P, Hawkesford M J, Varshney R K, Yang T, Nie C, Li Z, Ming B, Xiao Y, Xie Y, Li S. 2020. High-throughput estimation of crop traits:a review of ground and aerial phenotyping platforms. IEEE Geoscience and Remote Sensing Magazine, 9 (1):200-231.
doi: 10.1109/MGRS.2020.2998816 URL |
| [45] |
Kicherer A, Klodt M, Sharifzadeh S, Cremers D, Töpfer R, Herzog K. 2017. Automatic image-based determination of pruning mass as a determinant for yield potential in grapevine management and breeding. Australian Journal of Grape and Wine Research, 23:120-124.
doi: 10.1111/ajgw.12243 URL |
| [46] | Kicherer A, Roscher R, Herzog K, Šimon S, Förstner W, Töpfer R. 2013. Bat(berry analysis tool):a high-throughput image interpretation tool to acquire the number,diameter,and volume of grapevine berries. Vitis, 52:129-135. |
| [47] |
Klodt M, Herzog K, Topfer R, Cremers D. 2015. Field phenotyping of grapevine growth using dense stereo reconstruction. BMC Bioinformatics, 16:143.
doi: 10.1186/s12859-015-0560-x URL |
| [48] |
Klukas C, Chen D, Pape J M. 2014. Integrated analysis platform:an open-source information system for high-throughput plant phenotyping. Plant Physiology, 165:506-518.
doi: 10.1104/pp.113.233932 URL |
| [49] |
Kobayashi S, Goto-Yamamoto N, Hirochika H. 2004. Retrotransposon-induced mutations in grape skin color. Science, 304:982.
pmid: 15143274 |
| [50] |
Li M, Coneva V, Robbins K R, Clark D, Chitwood D, Frank M. 2021. Quantitative dissection of color patterning in the foliar ornamental coleus. Plant Physiol, 187:1310-1324.
doi: 10.1093/plphys/kiab393 URL |
| [51] |
Li M, Klein L L, Duncan K E, Jiang N, Chitwood D H, Londo J P, Miller A J, Topp C N. 2019. Characterizing 3d inflorescence architecture in grapevine using x-ray imaging and advanced morphometrics:implications for understanding cluster density. J Exp Bot, 70:6261-6276.
doi: 10.1093/jxb/erz394 URL |
| [52] |
Liang Z, Duan S, Sheng J, Zhu S, Ni X, Shao J, Liu C, Nick P, Du F, Fan P. 2019. Whole-genome resequencing of 472 Vitis accessions for grapevine diversity and demographic history analyses. Nature Communications, 10:1-12.
doi: 10.1038/s41467-018-07882-8 URL |
| [53] | Liu Chong-huai, Guo Jing-nan, Fan Xiu-cai. 2010. Some ampelometric leaf characteristics used in grape varieties identification. Sino-Overseas Grapevine & Wine,(1):31-34,36. (in Chinese) |
| 刘崇怀, 郭景南, 樊秀彩. 2010. 葡萄品种资源研究中的叶片测量性状. 中外葡萄与葡萄酒,(1):31-34,36. | |
| [54] | Liu Chong-huai, Kong Qing-shan, Guo Jing-nan, Pan Xing. 2003. Studies on the main fruit characters of grape germplasm reources. Chinese Agricultural Science Bulletin,(2):74-76,79. (in Chinese) |
| 刘崇怀, 孔庆山, 郭景南, 潘兴. 2003. 葡萄品种资源果实重要经济性状分析. 中国农学通报,(2):74-76,79. | |
| [55] | Liu Feng-zhi. 2017. Present situation and development trend of Chinese viticulture. Deciduous Fruits, 49:1-4. (in Chinese) |
| 刘凤之. 2017. 中国葡萄栽培现状与发展趋势. 落叶果树, 49:1-4. | |
| [56] | Liu S, Zeng X, Whitty M. 2020a. 3dbunch:A novel ios-smartphone application to evaluate the number of grape berries per bunch using image analysis techniques. IEEE Access, 8:114663-114674. |
| [57] | Liu S, Zeng X, Whitty M. 2020b. A vision-based robust grape berry counting algorithm for fast calibration-free bunch weight estimation in the field. Computers and Electronics in Agriculture, 173:105360. |
| [58] | Liu Xin-ming, Zhang Guo-hai, Liu Chong-huai, Guo Da-long, Fan Xiu-cai, Sun Hai-sheng. 2010. Evaluating on phenotypic diversity of grape leaves. Northern Horticulture,(6):1-4. (in Chinese) |
| 刘鑫铭, 张国海, 刘崇怀, 郭大龙, 樊秀彩, 孙海生. 2010. 葡萄叶片表型多样性分析. 北方园艺,(6):1-4. | |
| [59] | Liu Xun-ju, Zheng Zhi-xin, Jiu Song-tao, Xu Yan, Zhang Cai-xi. 2020. Application and prospect analysis of artificial intelligence in grape and wine industry. Sino-Overseas Grapevine & Wine,(5):44-48. (in Chinese) |
| 刘勋菊, 郑智心, 纠松涛, 徐岩, 张才喜. 2020. 人工智能在葡萄与葡萄酒产业中的应用及前景分析. 中外葡萄与葡萄酒,(5):44-48. | |
| [60] | Lopes C M, Torres A, Guzman R, Graça J, Reyes M, Vitorino G, Braga R, Monteiro A, Barriguinha A. 2017. Using an unmanned ground vehicle to scout vineyards for non-intrusive estimation of canopy features and grape yield// GiESCO International Meeting, 20th,Sustainable viticulture and wine making in climate chenge scenarios.17-21. |
| [61] | Lu Yanan, Zhuge Yaxian, Pei Qingyuan, Ma Wudan, Fan Xiucai, Liu Chonghuai, Guan Le, Fang Jinggui. 2019. Investigation and comprehensive evaluation on cluster shape of grape germplasm resources. Acta Horticulturae Sinica, 46 (8):1593-1603. (in Chinese) |
| 鲁雅楠, 诸葛雅贤, 裴清圆, 马吾丹, 樊秀彩, 刘崇怀, 管乐, 房经贵. 2019. 葡萄种质资源果穗穗形调查与综合评价. 园艺学报, 46 (8):1593-1603. | |
| [62] |
Louarn G, Lecoeur J, Lebon E. 2008. A three-dimensional statistical reconstruction model of grapevine(Vitis vinifera)simulating canopy structure variability within and between cultivar/training system pairs. Annals of Botany, 101:1167-1184.
doi: 10.1093/aob/mcm170 URL |
| [63] |
Mack J, Lenz C, Teutrine J, Steinhage V. 2017. High-precision 3D detection and reconstruction of grapes from laser range data for efficient phenotyping based on supervised learning. Computers and Electronics in Agriculture, 135:300-311.
doi: 10.1016/j.compag.2017.02.017 URL |
| [64] |
Mack J, Rist F, Herzog K, Töpfer R, Steinhage V. 2020. Constraint-based automated reconstruction of grape bunches from 3D range data for high-throughput phenotyping. Biosystems Engineering, 197:285-305.
doi: 10.1016/j.biosystemseng.2020.07.004 URL |
| [65] |
Marani R, Milella A, Petitti A, Reina G. 2021. Deep neural networks for grape bunch segmentation in natural images from a consumer-grade camera. Precision Agriculture, 22 (2):387-413.
doi: 10.1007/s11119-020-09736-0 URL |
| [66] |
Massonnet M, Cochetel N, Minio A, Vondras A M, Lin J, Muyle A, Garcia J F, Zhou Y, Delledonne M, Riaz S. 2020. The genetic basis of sex determination in grapes. Nature Communications, 11:1-12.
doi: 10.1038/s41467-019-13993-7 URL |
| [67] |
Miao C, Xu Y, Liu S, Schnable P S, Schnable J C. 2020. Increased power and accuracy of causal locus identification in time series genome-wide association in sorghum. Plant Physiol, 183:1898-1909.
doi: 10.1104/pp.20.00277 URL |
| [68] |
Milella A, Marani R, Petitti A, Reina G. 2019. In-field high throughput grapevine phenotyping with a consumer-grade depth camera. Computers and Electronics in Agriculture, 156:293-306.
doi: 10.1016/j.compag.2018.11.026 URL |
| [69] |
Min Z, Li R, Zhao X, Li R, Zhang Y, Liu M, Wei X, Fang Y, Chen S. 2018. Morphological variability in leaves of chinese wild Vitis species. Scientia Horticulturae, 238:138-146.
doi: 10.1016/j.scienta.2018.04.006 URL |
| [70] | Minervini M, Scharr H, Tsaftaris S A. 2015. Image analysis:the new bottleneck in plant phenotyping. IEEE Signal Processing Magazine, 32:126-131. |
| [71] |
Mir R R, Reynolds M, Pinto F, Khan M A, Bhat M A. 2019. High-throughput phenotyping for crop improvement in the genomics era. Plant Science, 282:60-72.
doi: 10.1016/j.plantsci.2019.01.007 URL |
| [72] |
Molitor D, Behr M, Hoffmann L, Evers D. 2012. Impact of grape cluster division on cluster morphology and bunch rot epidemic. American Journal of Enology Viticulture, 63:508-514.
doi: 10.5344/ajev.2012.12041 URL |
| [73] | Mu Jin-hu, Chen Yu-ze, Feng Hui, Li Wen-jian, Zhou Li-bin. 2016. A new revolution in crop breeding:the era of high-throughput phenomics. Plant Science Journal, 34:962-971. (in Chinese) |
| 穆金虎, 陈玉泽, 冯慧, 李文建, 周利斌. 2016. 作物育种学领域新的革命:高通量的表型组学时代. 植物科学学报, 34:962-971. | |
| [74] |
Ni X, Li C, Jiang H, Takeda F. 2020. Deep learning image segmentation and extraction of blueberry fruit traits associated with harvestability and yield. Horticulture Research, 7:110.
doi: 10.1038/s41438-020-0323-3 URL |
| [75] |
Muñoz-Espinoza C, Di Genova A, Sánchez A, Correa J, Espinoza A, Meneses C, Maass A, Orellana A, Hinrichsen P. 2020. Identification of snps and indels associated with berry size in table grapes integrating genetic and transcriptomic approaches. BMC Plant Biology, 20:1-21.
doi: 10.1186/s12870-019-2170-7 URL |
| [76] | Peng Yong-bin, Xie Xian-zhi. 2020. Application of phenomics in rice research. Chinese Journal of Rice Science, 34:300-306. (in Chinese) |
|
彭永彬, 谢先芝. 2020. 表型组学在水稻研究中的应用. 中国水稻科学, 34:300-306.
doi: 10.16819/j.1001-7216.2020.9083 |
|
| [77] |
Reshef N, Karn A, Manns D C, Mansfield A K, Cadle-Davidson L, Reisch B, Sacks G L. 2022. Stable QTL for malate levels in ripe fruit and their transferability across vitis species. Horticulture Research, 9:uhac009.
doi: 10.1093/hr/uhac009 URL |
| [78] | Reynolds M, Chapman S, Crespo-Herrera L, Molero G, Mondal S, Pequeno D N L, Pinto F, Pinera-Chavez F J, Poland J, Rivera-Amado C, Saint Pierre C, Sukumaran S. 2020. Breeder friendly phenotyping. Plant Science, 295:110396. |
| [79] |
Rist F, Gabriel D, Mack J, Steinhage V, Töpfer R, Herzog K. 2019. Combination of an automated 3D field phenotyping workflow and predictive modelling for high-throughput and non-invasive phenotyping of grape bunches. Remote Sensing, 11 (24):2953.
doi: 10.3390/rs11242953 URL |
| [80] |
Rist F, Herzog K, Mack J, Richter R, Steinhage V, Topfer R. 2018. High-precision phenotyping of grape bunch architecture using fast 3D sensor and automation. Sensors, 18 (3):763.
doi: 10.3390/s18030763 URL |
| [81] |
Rodríguez-Pulido F J, Barbin D F, Sun D-W, Gordillo B, González-Miret M L, Heredia F J. 2013. Grape seed characterization by nir hyperspectral imaging. Postharvest Biology and Technology, 76:74-82.
doi: 10.1016/j.postharvbio.2012.09.007 URL |
| [82] | Santos T T, de Souza L L, dos Santos A A, Avila S. 2020. Grape detection,segmentation,and tracking using deep neural networks and three-dimensional association. Computers and Electronics in Agriculture, 170:105247. |
| [83] |
Sarigu M, Grillo O, Lo Bianco M, Ucchesu M, d'Hallewin G, Loi M C, Venora G, Bacchetta G. 2017. Phenotypic identification of plum varieties (Prunus domestica L.)by endocarps morpho-colorimetric and textural descriptors. Computers and Electronics in Agriculture, 136:25-30.
doi: 10.1016/j.compag.2017.02.009 URL |
| [84] | Schneider T, Paulus G, Anders K-H. 2020. Towards predicting vine yield:conceptualization of 3D grape models and derivation of reliable physical and morphological parameters. GI_Forum, 1:73-88. |
| [85] |
Schöler F, Steinhage V. 2015. Automated 3d reconstruction of grape cluster architecture from sensor data for efficient phenotyping. Computers and Electronics in Agriculture, 114:163-177.
doi: 10.1016/j.compag.2015.04.001 URL |
| [86] | Seethepalli A, Guo H, Liu X, Griffiths M, Almtarfi H, Li Z, Liu S, Zare A, Fritschi F B, Blancaflor E B, Ma X F, York L M. 2020. Rhizovision crown:An integrated hardware and software platform for root crown phenotyping, Plant Phenomics, 2020:3074916. |
| [87] |
Snouffer A, Kraus C, van der Knaap E. 2020. The shape of things to come:ovate family proteins regulate plant organ shape. Curr Opin Plant Biol, 53:98-105.
doi: S1369-5266(19)30096-2 pmid: 31837627 |
| [88] | Sozzi M, Kayad A, Tomasi D, Lovat L, Marinello F, Sartori L. 2019. Assessment of grapevine yield and quality using a canopy spectral index in white grape variety//Precision agriculture’19. Wageningen Academic Publishers:111-129. |
| [89] | Tang Z, Yang J, Li Z, Qi F. 2020. Grape disease image classification based on lightweight convolution neural networks and channelwise attention. Computers and Electronics in Agriculture, 178:105735. |
| [90] |
Tello J, Cubero S, Blasco J, Tardaguila J, Aleixos N, Ibanez J. 2016. Application of 2D and 3D image technologies to characterise morphological attributes of grapevine clusters. J Sci Food Agric, 96:4575-4583.
doi: 10.1002/jsfa.7675 URL |
| [91] | Tello J, Forneck A. 2018. A double-sigmoid model for grapevine bunch compactness development. OENO One, 52:307-316. |
| [92] |
Tello J, Montemayor M I, Forneck A, Ibanez J. 2018. A new image-based tool for the high throughput phenotyping of pollen viability:Evaluation of inter- and intra-cultivar diversity in grapevine. Plant Methods, 14:3.
doi: 10.1186/s13007-017-0267-2 URL |
| [93] |
This P, Lacombe T, Thomas M R. 2006. Historical origins and genetic diversity of wine grapes. Trends Genet, 22:511-519.
doi: 10.1016/j.tig.2006.07.008 URL |
| [94] |
Underhill A, Hirsch C, Clark M. 2020a. Image-based phenotyping identifies quantitative trait loci for cluster compactness in grape. Journal of the American Society for Horticultural Science, 145:363-373.
doi: 10.21273/JASHS04932-20 URL |
| [95] | Underhill A N, Hirsch C D, Clark M D. 2020b. Evaluating and mapping grape color using image-based phenotyping. Plant Phenomics,8086309. |
| [96] | Victorino G, Maia G, Queiroz J, Braga R, Marques J, Santos-Victor J, Lopes C. 2019. Grapevine yield prediction using image analysis-improving the estimation of non-visible bunches,12th EFITA International Conference; Rhodes Island,Greece:60-65. |
| [97] |
Vondras A M, Minio A, Blanco-Ulate B, Figueroa-Balderas R, Penn M A, Zhou Y, Seymour D, Ye Z, Liang D, Espinoza L K. 2019. The genomic diversification of grapevine clones. BMC Genomics, 20:1-19.
doi: 10.1186/s12864-018-5379-1 URL |
| [98] |
Walker A R, Lee E, Bogs J, McDavid D A, Thomas M R, Robinson S P. 2007. White grapes arose through the mutation of two similar and adjacent regulatory genes. The Plant Journal, 49:772-785.
doi: 10.1111/j.1365-313X.2006.02997.x URL |
| [99] | Wang Haomiao, Song Miaoyu, Li Xiang, Hu Chaoyang, Lu Renxiang, Wang Xiang, Ma Huiqin. 2021. High efficient grapevine growth monitor and in-lane deficiency localization by UAV hyperspectral remote sensing. Acta Horticulturae Sinica, 48 (8):1626-1634. |
| 王浩淼, 宋苗语, 李翔, 扈朝阳, 鲁任翔, 王翔, 马会勤. 2021. 无人机高光谱遥感监测葡萄长势与缺株定位. 园艺学报, 48 (8):1626-1634. | |
| [100] |
Wang Y, Wen W, Wu S, Wang C, Yu Z, Guo X, Zhao C. 2019. Maize plant phenotyping:comparing 3D laser scanning,multi-view stereo reconstruction,and 3D digitizing estimates. Remote Sensing, 11:63.
doi: 10.3390/rs11010063 URL |
| [101] | Wang Yong-jian, Wen Wei-liang, Guo Xin-yu, Zhao Chun-jiang. 2014. Three-dimensional reconstruction of plant leaf blade based on point cloud data. Journal of Agricultural Science and Technology, 16 (5):83-89. (in Chinese) |
| 王勇健, 温维亮, 郭新宇, 赵春江. 2014. 基于点云数据的植物叶片三维重建. 中国农业科技导报, 16 (5):83-89. | |
| [102] | Wang Z, Chai F, Zhu Z, Elias G K, Xin H, Liang Z, Li S. 2020. The inheritance of cold tolerance in seven interspecific grape populations. Scientia Horticulturae, 266:109260. |
| [103] |
Washburn J D, Burch M B, Valdes Franco J A. 2019. Predictive breeding for maize:making use of molecular phenotypes,machine learning,and physiological crop models. Crop Science, 60 (2):622-638.
doi: 10.1002/csc2.20052 URL |
| [104] | Watt M, Fiorani F, Usadel B, Rascher U, Muller O, Schurr U. 2020. Phenotyping:new windows into the plant for breeders. Annual Review of Plant Biology, 71:annurev-arplant-042916-041124. |
| [105] |
Wolkovich E M, García de Cortázar-Atauri I, Morales-Castilla I, Nicholas K A, Lacombe T. 2018. From pinot to xinomavro in the world's future wine-growing regions. Nature Climate Change, 8:29-37.
doi: 10.1038/s41558-017-0016-6 URL |
| [106] |
Wu S, Wen W, Wang Y, Fan J, Wang C, Gou W, Guo X. 2020. Mvs-pheno:a portable and low-cost phenotyping platform for maize shoots using multiview stereo 3d reconstruction. Plant Phenomics,Doi: 10.1080/01603477.2020.1848437.
doi: 10.1080/01603477.2020.1848437 |
| [107] |
Wycislo A P, Clark J R, Karcher D E. 2008. Fruit shape analysis of vitis using digital photography. HortScience, 43:677-680.
doi: 10.21273/HORTSCI.43.3.677 URL |
| [108] |
Xin B, Liu S, Whitty M. 2020. Three‐dimensional reconstruction of Vitis vinifera(L.)cvs pinot noir and merlot grape bunch frameworks using a restricted reconstruction grammar based on the stochastic l‐system. Australian Journal of Grape and Wine Research, 26 (3):207-219.
doi: 10.1111/ajgw.12444 URL |
| [109] | Xu Ling-xiang, Chen Jia-wei, Ding Guo-hui, Lu Wei, Ding Yan-feng, Zhu Yan, Zhou Ji. 2020. Indoor phenotyping platforms and associated trait measurement:progress and prospects. Smart Agriculture,(2):23-42. (in Chinese) |
| 徐凌翔, 陈佳玮, 丁国辉, 卢伟, 丁艳锋, 朱艳, 周济. 2020. 室内植物表型平台及性状鉴定研究进展和展望. 智慧农业,(2):23-42. | |
| [110] | Yang S, Fresnedo-Ramírez J, Wang M, Cote L, Schweitzer P, Barba P, Takacs E M, Clark M, Luby J, Manns D C. 2016. A next-generation marker genotyping platform(AmpSeq)in heterozygous crops:a case study for marker-assisted selection in grapevine. Horticulture Research, 3:1-12. |
| [111] |
Yang W, Feng H, Zhang X, Zhang J, Doonan J H, Batchelor W D, Xiong L, Yan J. 2020. Crop phenomics and high-throughput phenotyping:past decades,current challenges,and future perspectives. Molecular Plant, 13:187-214.
doi: 10.1016/j.molp.2020.01.008 URL |
| [112] |
Yang W, Guo Z, Huang C, Zhang J, Doonan J H, Batchelor W D, Xiong L, Yan J. 2014. Combining high-throughput phenotyping and genome-wide association studies to reveal natural genetic variation in rice. Nat Commun, 5:5087.
doi: 10.1038/ncomms6087 URL |
| [113] | Yang Xin, Sun Yu-jie, Zhou Bi-jiang, Fu Wen-jing, Fang Yu-lin, Meng Jiang-fei. 2021. Evaluation and analysis of leaf morphology of grape varieties cultivated in China. Northern Horticulture,(1):23-29. (in Chinese) |
| 杨鑫, 孙宇杰, 周碧江, 付文静, 房玉林, 孟江飞. 2021. 中国现有葡萄品种叶片形态评价与分析. 北方园艺,(1):23-29. | |
| [114] | Zhang Heng, Liu Zhong-jie, Fan Xiu-cai, Zhang Chuan, Cui Li-wen, Liu Chong-huai, Fang Jing-gui. 2017. Genome-wide association mapping of berry shape traits via the reduced representation sequencing in grape. Acta Horticulturae Sinica, 44 (10):1959-1968. (in Chinese) |
| 张恒, 刘众杰, 樊秀彩, 张川, 崔力文, 刘崇怀, 房经贵. 2017. 葡萄果粒形状简化基因组关联分析. 园艺学报, 44 (10):1959-1968. | |
| [115] | Zhang Peian, Fan Xiucai, Liu Zhongjie, Wu Weiming, Liu Chonghuai, Fang Jinggui. 2018. Investigation and analysis on the berry shape of grape germplasm resources. Acta Horticulturae Sinica, 45 (8):1456-1466. (in Chinese) |
| 张培安, 樊秀彩, 刘众杰, 吴伟民, 刘崇怀, 房经贵. 2018. 葡萄种质资源果形性状的分析. 园艺学报, 45 (8):1456-1466. | |
| [116] | Zhang Pei-an, Zhang Wen-ying, Jiu Song-tao, Zhang Ke-kun, Zhang Chao-bo, Fang Jing-gui, Liu Chong-huai. 2017. Analyses on pericarp color and fruit coloring characters of grape(Vitis spp.). Journal of Plant Resources and Environment, 26:8-17. (in Chinese) |
| 张培安, 张文颖, 纠松涛, 张克坤, 张超博, 房经贵, 刘崇怀. 2017. 葡萄(Vitis spp.)果皮颜色及果实着色性状分析. 植物资源与环境学报, 26:8-17. | |
| [117] |
Zhang X, Huang C, Wu D, Qiao F, Li W, Duan L, Wang K, Xiao Y, Chen G, Liu Q, Xiong L, Yang W, Yan J. 2017. High-throughput phenotyping and QTL mapping reveals the genetic architecture of maize plant growth. Plant Physiology, 173:1554-1564.
doi: 10.1104/pp.16.01516 URL |
| [118] |
Zhao C, Zhang Y, Du J, Guo X, Wen W, Gu S, Wang J, Fan J. 2019. Crop phenomics:current status and perspectives. Frontiers in Plant Science, 10:714.
doi: 10.3389/fpls.2019.00714 URL |
| [119] | Zhao Chun-jiang. 2019. State-of-the-art and recommended developmental strategic objectives of smart agriculture. Smart Agriculture,(1):1-7. (in Chinese) |
| 赵春江. 2019. 智慧农业发展现状及战略目标研究. 智慧农业(中英文),(1):1-7. | |
| [120] | Zhou Ji, Cheng Tao, Zhu Yan, Wang Xiu-e. 2018. Plant phenomics:history,present status and challenges. Journal of Nanjing Agricultural University, 41:580-588. (in Chinese) |
| 周济, 程涛, 朱艳, 王秀娥. 2018. 植物表型组学:发展、现状与挑战. 南京农业大学学报, 41:580-588. | |
| [121] | Zhou Y, Massonnet M, Sanjak J S, Cantu D, Gaut B S. 2017. Evolutionary genomics of grape(Vitis vinifera ssp. vinifera)domestication. Proceedings of the National Academy of Sciences, 114:11715-11720. |
| [122] |
Zhou Y, Minio A, Massonnet M, Solares E, Lv Y, Beridze T, Cantu D, Gaut B S. 2019. The population genetics of structural variants in grapevine domestication. Nature Plants, 5:965-979.
doi: 10.1038/s41477-019-0507-8 URL |
| [123] |
Zhu J, Dai Z, Vivin P, Gambetta G A, Henke M, Peccoux A, Ollat N, Delrot S. 2018. A 3-D functional-structural grapevine model that couples the dynamics of water transport with leaf gas exchange. Annals of Botany, 121:833-848.
doi: 10.1093/aob/mcx141 URL |
| [124] |
Zhu J, Génard M, Poni S, Gambetta G A, Vivin P, Vercambre G, Trought M C T, Ollat N, Delrot S, Dai Z. 2019. Modelling grape growth in relation to whole-plant carbon and water fluxes. Journal of Experimental Botany, 70:2505-2521.
doi: 10.1093/jxb/ery367 URL |
| [1] | WANG Xiaochen, NIE Ziye, LIU Xianju, DUAN Wei, FAN Peige, and LIANG Zhenchang, . Effects of Abscisic Acid on Monoterpene Synthesis in‘Jingxiangyu’Grape Berries [J]. Acta Horticulturae Sinica, 2023, 50(2): 237-249. |
| [2] | WANG Baoliang, LIU Fengzhi, JI Xiaohao, WANG Xiaodi, SHI Xiangbin, ZHANG Yican, LI Peng, and WANG Haibo. A New Early Ripening Grape Cultivar‘Huapu Zaoyu’for Table [J]. Acta Horticulturae Sinica, 2022, 49(S2): 33-34. |
| [3] | WANG Baoliang, WANG Haibo, JI Xiaohao, WANG Xiaodi, SHI Xiangbin, WANG Zhiqiang, WANG Xiaolong, and LIU Fengzhi. A New Middle Ripening Grape Cultivar‘Huapu Huangyu’for Table [J]. Acta Horticulturae Sinica, 2022, 49(S2): 35-36. |
| [4] | A Late-maturing Seedless Grape Cultivar‘Zilongzhu’. A Late-maturing Seedless Grape Cultivar‘Zilongzhu’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 37-38. |
| [5] | SHI Xiaoxin, DU Guoqiang, YANG Lili, QIAO Yuelian, HUANG Chengli, WANG Suyue, ZHAO Yuexin, WEI Xiaohui, WANG Li, and QI Xiangli. A Late-ripening Seedless Grape Cultivar‘Hongfeng Wuhe’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 39-40. |
| [6] | WU Yueyan, CHEN Tianchi, WANG Liru, HAN Shanqi, and FU Tao. A New Table Grape Cultivar‘Yongzaohong’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 41-42. |
| [7] | DENG Chaojun, XU Qizhi, JIANG Jimou, HU Wenshun, ZHENG Shaoquan, CHEN Xiuping, JIANG Fan, XU Jiahui, SU Wenbing, ZHANG Yaling, and HUANG Jingfeng. A New Longan Cultivar‘Chunxiang’with Rich Aroma [J]. Acta Horticulturae Sinica, 2022, 49(S2): 75-76. |
| [8] | DENG Chaojun, CHEN Xiuping, XU Qizhi, JIANG Jimou, ZHENG Shaoquan, HU Wenshun, JIANG Fan, XU Jiahui, SU Wenbing, ZHANG Yaling, and HUANG Jingfeng. A New Longan Cultivar‘Fuxiang’with Rich Aroma [J]. Acta Horticulturae Sinica, 2022, 49(S2): 77-78. |
| [9] | WANG Xiaoyue, YAN Ailing, ZHANG Guojun, WANG Huiling, REN Jiancheng, LIU Zhenhua, SUN Lei, and XU Haiying, . A New Grape Cultivar‘Ruidu Wanhong’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 29-30. |
| [10] | WANG Yulin, ZHENG Yunfeng, TAN Baishen, CHEN Huiping, and SUN Qingming, . A New Pitaya Cultivar‘Daqiu 4’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 59-60. |
| [11] | XUE Chenchen, HUANG Lu, ZHANG Xiaoyan, SU Xiaoming, SU Ruocheng, YUAN Xingxing, and CHEN Xin. A New Vegetable Soybean Cultivar‘Sucheng 4’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 89-90. |
| [12] | CHEN Meiyuan, LIAO Jianhua, CAI Zhixin, GUO Zhongjie, LU Yuanping, and WANG Zesheng. A New Agaricus bisporus Cultivar‘Fumo 52’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 97-100. |
| [13] | WANG Xiaobin, ZHANG Dong, SHI Xiaohua, LI Danqing, ZHANG Runlong, SHAO Lingmei, XU Tong, XIA Yiping, and ZHANG Jiaping, . A New Paeonia lactiflora Cultivar‘Purple Heart’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 115-116. |
| [14] | LUO Qing, DING Changchun, XIE Zhenxing, HUANG Xinyi, QIN Qian, and LU Zuzheng. A New Cymbidium Cultivar‘Yellow Sky’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 129-130. |
| [15] | YI Shuangshuang, LIAO Yi, HUANG Mingzhong, Li Chonghui, ZHANG Zhiqun, and LU Shunjiao, . A New Dendrobium Cultivar‘Xiaotaohong’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 135-136. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd