
Acta Horticulturae Sinica ›› 2022, Vol. 49 ›› Issue (7): 1441-1457.doi: 10.16420/j.issn.0513-353x.2021-0553
• Research Papers • Previous Articles Next Articles
ZHENG Lin, WANG Shuai, LIU Yunuo, DU Meixia, PENG Aihong, HE Yongrui, CHEN Shanchun( ), ZOU Xiuping(
), ZOU Xiuping( )
)
Received:2022-02-08
Revised:2022-04-25
Online:2022-07-25
Published:2022-07-29
Contact:
CHEN Shanchun,ZOU Xiuping
E-mail:chenshanchun@cric.cn;zouxiuping@cric.cn
CLC Number:
ZHENG Lin, WANG Shuai, LIU Yunuo, DU Meixia, PENG Aihong, HE Yongrui, CHEN Shanchun, ZOU Xiuping. Gene Cloning and Expression Analysis of NAC Gene in Citrus in Response to Huanglongbing[J]. Acta Horticulturae Sinica, 2022, 49(7): 1441-1457.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2021-0553
| 用途Amplification | 名称Name | 序列(5′-3′)Sequence |
|---|---|---|
| 黄龙病常规 PCR 检测 Huanglongbing routine PCR test | OI1 OI2C | F:GCGCGTATGCAATACGAGCGGCA |
| R:GCCTCGCGACTTCGCAACCCAT | ||
| 基因克隆Gene cloning | CsNAC21/22 | F:CGCGGATCCATGGGGCTAAGAGATATTGGAGCTAC |
| R:CCGGAATTCTCATAGGAAAACCATGCTGTTATCCAAT | ||
| CsNAC68 | F:TCCCCCGGGATGCCCAAAGAGGACCGAG | |
| R:GCGTCGACTCAATGAAGATACGCCATT | ||
| CsNAC78 | F:CGCGGATCCATGGACTATAATTGCCTGGGATACAG | |
| R:CCGGAATTCCTATATGAAGAAAAACAAGTTGTATAGAACAGAAG | ||
| 亚细胞定位Subcellular localization | CsNAC21/22 | F:GGGGTACCATGGGGCTAAGAGATATTGG |
| R:ACGCGTCGACTAGGAAAACCATGCTGTTAT | ||
| CsNAC68 | F:GGGGTACCATGCCCAAAGAGGACCGAGT | |
| R:ACGCGTCGACATGAAGATACGCCATTTCCC | ||
| CsNAC78 | F:GGGGTACCATGGACTATAATTGCCTGGG | |
| R:ACGCGTCGACTATGAAGAAAAACAAGTTGT |
Table 1 The sequences of common PCR primer pairs
| 用途Amplification | 名称Name | 序列(5′-3′)Sequence |
|---|---|---|
| 黄龙病常规 PCR 检测 Huanglongbing routine PCR test | OI1 OI2C | F:GCGCGTATGCAATACGAGCGGCA |
| R:GCCTCGCGACTTCGCAACCCAT | ||
| 基因克隆Gene cloning | CsNAC21/22 | F:CGCGGATCCATGGGGCTAAGAGATATTGGAGCTAC |
| R:CCGGAATTCTCATAGGAAAACCATGCTGTTATCCAAT | ||
| CsNAC68 | F:TCCCCCGGGATGCCCAAAGAGGACCGAG | |
| R:GCGTCGACTCAATGAAGATACGCCATT | ||
| CsNAC78 | F:CGCGGATCCATGGACTATAATTGCCTGGGATACAG | |
| R:CCGGAATTCCTATATGAAGAAAAACAAGTTGTATAGAACAGAAG | ||
| 亚细胞定位Subcellular localization | CsNAC21/22 | F:GGGGTACCATGGGGCTAAGAGATATTGG |
| R:ACGCGTCGACTAGGAAAACCATGCTGTTAT | ||
| CsNAC68 | F:GGGGTACCATGCCCAAAGAGGACCGAGT | |
| R:ACGCGTCGACATGAAGATACGCCATTTCCC | ||
| CsNAC78 | F:GGGGTACCATGGACTATAATTGCCTGGG | |
| R:ACGCGTCGACTATGAAGAAAAACAAGTTGT |
| 目的基因 Target gene | 引物序列(5′-3′) Primer sequence |
|---|---|
| Cs1g09640.1 | F:AGCCATGGGACTTACCAACG;R:ACCAGTAAGGCCGTTTCCAG |
| Cs4g12500.1 | F:GATCATCAGCTTCCGGGGTT;R:TTCGATGCTGAGCGGTTTCT |
| Cs5g29650.1(CsNAC21/22) | F:GGTGGCAAAGCTCAATGCAA;R:GCGTCTTCCTCATCCCTACG |
| Cs6g14200.1 | F:AAGGGAAGTGCCTCAACGAG;R:TCGTCTCCGGAGGATGAACT |
| Cs8g14700.1 | F:GTCAAGGAGACACCGAGCAA;R:AGCCTGTGGCCTTCCAATAC |
| orange1.1t00587.1 | F:AGGAATTTGTCCTCCTTCGTT;R:AGCCGAAGGTTGAGGAGTTG |
| orange1.1t00588.1 | F:TACAAAGGTCGTGGCTCCAG;R:TGCAGCTCGGGTATAGTCCT |
| orange1.1t00589.1(CsNAC68) | F:CGAAGGAATTTGTCCTCTTCCG;R:AGACCGCGGTTTATCTGTGG |
| orange1.1t00590.1(CsNAC78) | F:ACTGGTGGCGATCGTCAAAT;R:AGAACAGAAGAGATTGCCTGATGA |
Table 2 The sequences of qRT-PCR primer pairs
| 目的基因 Target gene | 引物序列(5′-3′) Primer sequence |
|---|---|
| Cs1g09640.1 | F:AGCCATGGGACTTACCAACG;R:ACCAGTAAGGCCGTTTCCAG |
| Cs4g12500.1 | F:GATCATCAGCTTCCGGGGTT;R:TTCGATGCTGAGCGGTTTCT |
| Cs5g29650.1(CsNAC21/22) | F:GGTGGCAAAGCTCAATGCAA;R:GCGTCTTCCTCATCCCTACG |
| Cs6g14200.1 | F:AAGGGAAGTGCCTCAACGAG;R:TCGTCTCCGGAGGATGAACT |
| Cs8g14700.1 | F:GTCAAGGAGACACCGAGCAA;R:AGCCTGTGGCCTTCCAATAC |
| orange1.1t00587.1 | F:AGGAATTTGTCCTCCTTCGTT;R:AGCCGAAGGTTGAGGAGTTG |
| orange1.1t00588.1 | F:TACAAAGGTCGTGGCTCCAG;R:TGCAGCTCGGGTATAGTCCT |
| orange1.1t00589.1(CsNAC68) | F:CGAAGGAATTTGTCCTCTTCCG;R:AGACCGCGGTTTATCTGTGG |
| orange1.1t00590.1(CsNAC78) | F:ACTGGTGGCGATCGTCAAAT;R:AGAACAGAAGAGATTGCCTGATGA |
| 基因ID Gene ID | 基因 Gene | CLas vs. 空白对照 CLas vs. Mock | |
|---|---|---|---|
| 叶片中脉Midrib | 根Root | ||
| Cs1g09640.1 | CsNTL9-1 | 1.252 | 4.283 |
| Cs4g12500.1 | CsNAC | — | — |
| Cs5g29650.1 | CsNAC21/22 | — | — |
| Cs6g14240.1 | CsNAC090-1 | 5.469 | — |
| Cs8g14700.1 | CsNAC090-2 | -1.172 | — |
| orange1.1t00587.1 | CsNAC050 | 2.315 | 5.735 |
| orange1.1t00588.1 | CsNTL9-2 | — | 20.474 |
| orange1.1t00589.1 | CsNAC68 | — | 4.163 |
| orange1.1t00590.1 | CsNAC78 | 1.633 | 11.316 |
Table 3 Comparative analysis of CsNAC family transcriptome in response to CLas infection in Jincheng
| 基因ID Gene ID | 基因 Gene | CLas vs. 空白对照 CLas vs. Mock | |
|---|---|---|---|
| 叶片中脉Midrib | 根Root | ||
| Cs1g09640.1 | CsNTL9-1 | 1.252 | 4.283 |
| Cs4g12500.1 | CsNAC | — | — |
| Cs5g29650.1 | CsNAC21/22 | — | — |
| Cs6g14240.1 | CsNAC090-1 | 5.469 | — |
| Cs8g14700.1 | CsNAC090-2 | -1.172 | — |
| orange1.1t00587.1 | CsNAC050 | 2.315 | 5.735 |
| orange1.1t00588.1 | CsNTL9-2 | — | 20.474 |
| orange1.1t00589.1 | CsNAC68 | — | 4.163 |
| orange1.1t00590.1 | CsNAC78 | 1.633 | 11.316 |
| 基因 Gene | 根 Root | 叶脉 Midrib | 基因 Gene | 根 Root | 叶脉 Midrib | |
|---|---|---|---|---|---|---|
| CsNTL9-1 | 2.42 ± 0.31 | 1.16 ± 0.24 | CsNAC050 | 4.24 ± 0.37 | 2.63 ± 0.44 | |
| CsNAC | 0.93 ± 0.09 | 2.01 ± 0.32 | CsNTL9-2 | 1.49 ± 0.15 | 1.68 ± 0.18 | |
| CsNAC21/22 | 0.49 ± 0.13 | 0.68 ± 0.10 | CsNAC68 | 8.62 ± 0.39 | 2.32 ± 0.18 | |
| CsNAC090-1 | 0.75 ± 0.12 | 1.38 ± 0.22 | CsNAC78 | 6.62 ± 0.29 | 1.86 ± 0.04 | |
| CsNAC090-2 | 0.97 ± 0.04 | 0.63 ± 0.10 |
Table 4 Relative expression levels of 9 NAC genes in Jincheng induced by HLB
| 基因 Gene | 根 Root | 叶脉 Midrib | 基因 Gene | 根 Root | 叶脉 Midrib | |
|---|---|---|---|---|---|---|
| CsNTL9-1 | 2.42 ± 0.31 | 1.16 ± 0.24 | CsNAC050 | 4.24 ± 0.37 | 2.63 ± 0.44 | |
| CsNAC | 0.93 ± 0.09 | 2.01 ± 0.32 | CsNTL9-2 | 1.49 ± 0.15 | 1.68 ± 0.18 | |
| CsNAC21/22 | 0.49 ± 0.13 | 0.68 ± 0.10 | CsNAC68 | 8.62 ± 0.39 | 2.32 ± 0.18 | |
| CsNAC090-1 | 0.75 ± 0.12 | 1.38 ± 0.22 | CsNAC78 | 6.62 ± 0.29 | 1.86 ± 0.04 | |
| CsNAC090-2 | 0.97 ± 0.04 | 0.63 ± 0.10 |
| 名称 Name | CAP号 CAP ID | ORF/bp | 氨基酸数 Number of amino acid | 分子量 Molecular weight | 等电点 pI | 亲水性平均值 Grand average of hydropathicity | 脂肪指数 Aliphatic index | 不稳定系数 Instability index | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|---|---|
| CsNAC21/22 | Cs5g29650.1 | 921 | 306 | 35 107.14 | 5.43 | -0.712 | 66.84 | 43.83 | 细胞核 Nuclear |
| CsNAC68 | orange1.1t00589.1 | 1 512 | 503 | 56 809.43 | 6.59 | -0.730 | 58.99 | 41.30 | 细胞核 Nuclear |
| CsNAC78 | orange1.1t00590.1 | 459 | 152 | 17 691.99 | 8.39 | -0.543 | 71.84 | 32.27 | 细胞质 Cytoplasm |
Table 5 Amino acid sequence analysis of three NAC genes in citrus
| 名称 Name | CAP号 CAP ID | ORF/bp | 氨基酸数 Number of amino acid | 分子量 Molecular weight | 等电点 pI | 亲水性平均值 Grand average of hydropathicity | 脂肪指数 Aliphatic index | 不稳定系数 Instability index | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|---|---|
| CsNAC21/22 | Cs5g29650.1 | 921 | 306 | 35 107.14 | 5.43 | -0.712 | 66.84 | 43.83 | 细胞核 Nuclear |
| CsNAC68 | orange1.1t00589.1 | 1 512 | 503 | 56 809.43 | 6.59 | -0.730 | 58.99 | 41.30 | 细胞核 Nuclear |
| CsNAC78 | orange1.1t00590.1 | 459 | 152 | 17 691.99 | 8.39 | -0.543 | 71.84 | 32.27 | 细胞质 Cytoplasm |
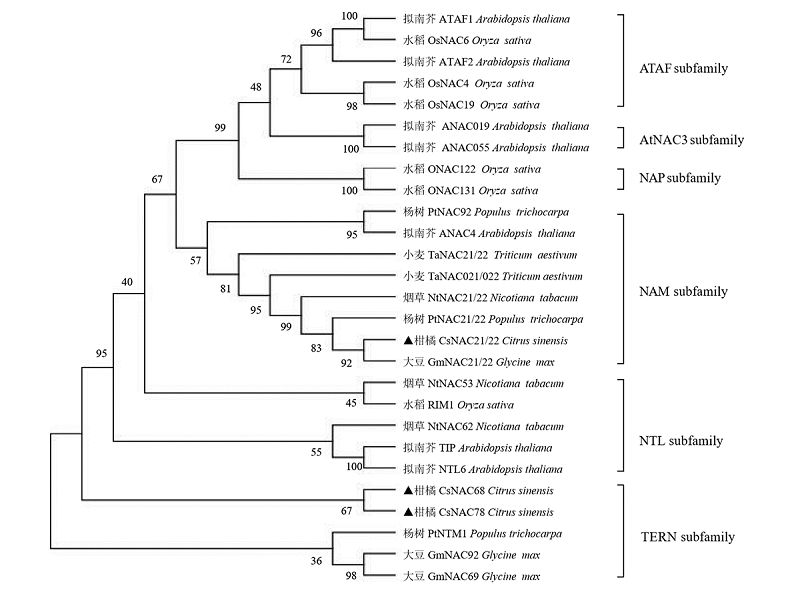
Fig. 3 CsNAC phylogenetic tree analysisConstructed by Neighbor-Joining method with the full-length sequence of the protein,and the Bootstrap value of 1 000 copies is indicated on each node,and the number on the branch is the self-spreading value(%).
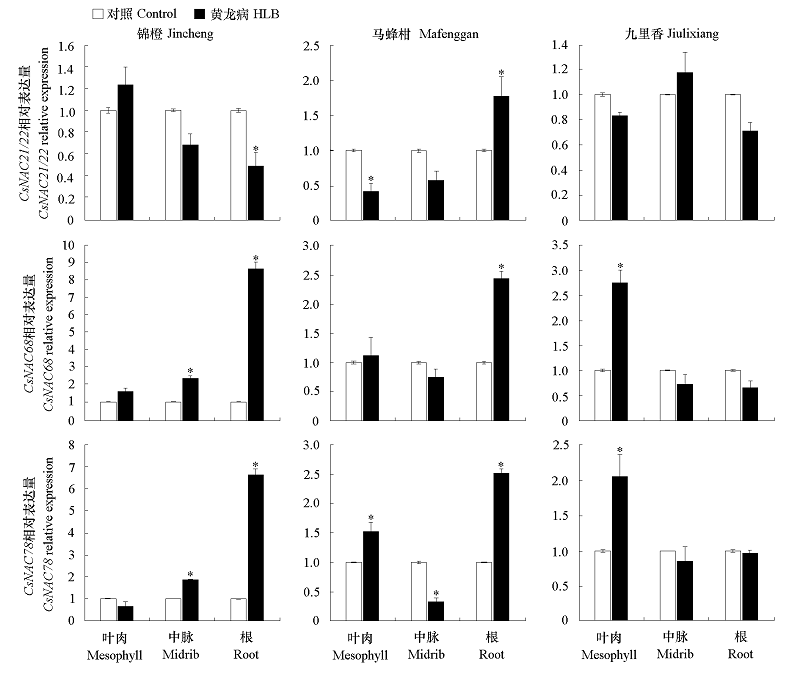
Fig. 5 Analysis of the relative expression levels of three NAC genes in different tissues of Jincheng,Mafenggan and Jiulixiang“*”Represents the significance of the difference with healthy plants of the same cultivar(P < 0.05).
| [1] |
Aida M, Tshida T, Fukaki H, Fujisawa H, Tasaka M. 1997. Genes involved in organ separation in Arabidopsis:an analysis of the cup-shaped cotyledon mutant. Plant Cell, 9:841-857.
pmid: 9212461 |
| [2] |
Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, De Castro E, Duvaud S, Flegel V, Fortier A, Gasteiger E, Grosdidier A, Hernandez C, Ioannidis V, Kuznetsov d, Liechti r, Moretti S, Mostaguir K, Redaschi N, Rossier G, Xenarios I, Stockinger H. 2012. ExPASy:SIB bioinformatics resource portal. Nucleic Acids Research, 40 (W1):W597-603.
doi: 10.1093/nar/gks400 URL |
| [3] |
Baldwin E, Plotto A, Bai J, Manthey J, Zhao W, Raithore S, Trey M. 2018. Effect of abscission zone formation on orange(Citrus sinensis) fruit/juice quality for trees affected by Huanglongbing(HLB). Journal of Agricultural and Food Chemistry, 66 (11):2877-2890.
doi: 10.1021/acs.jafc.7b05635 URL |
| [4] | Bove J M. 2006. Huanglongbing:a destructive,newly-emerging,century-old disease of citrus. Journal of Plant Pathology, 88 (1):7-37. |
| [5] |
Bu Q Y, Jiang H L, Li C B, Zhai Q Z, Zhang J, Wu X Y, Sun J Q, Xie Q, Li C Y. 2008. Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Research, 18 (7):756-767.
doi: 10.1038/cr.2008.53 URL |
| [6] |
Chen N, Wu S H, Fu J L, Cao B H, Lei J J, Chen C M, Jiang J. 2016. Overexpression of the eggplant(Solanum melongena)NAC family transcription factor SmNAC suppresses resistance to bacterial wilt. Scientific Reports, 6 (1):1-20.
doi: 10.1038/s41598-016-0001-8 URL |
| [7] | Chen Qian, You Shuang-mei, Xing Le-hua, Xu Fan, Luo Ming, Guo Qi-gao. 2021. Research progress of NAC transcription factors in fruit trees. Molecular Plant Breeding, 19 (19):6396-6405. (in Chinese) |
| 陈倩, 游双梅, 邢乐华, 徐凡, 罗明, 郭启高. 2021. 果树NAC转录因子的研究进展. 分子植物育种, 19 (19):6396-6405. | |
| [8] |
Chen Y J, Perera V, Christiansen M W, Holme I B, Gregersen P L, Grant M R, Collinge D B, Lyngkjar M F. 2013. The barley HvNAC6 transcription factor affects ABA accumulation and promotes basal resistance against powdery mildew. Plant Molecular Biology, 83 (6):577-590.
doi: 10.1007/s11103-013-0109-1 URL |
| [9] | Cheng Bao-ping, Zhao Hong-wei, Peng Ai-tian, Song Xiao-bing, Ling Jin-feng, Chen Xia. 2016. Investigation on the occurrence of citrus psyllid,a transmission agent of citrus greening disease in Guangdong orchards. Plant Protection, 42 (1):189-192,196. (in Chinese) |
| 程保平, 赵弘巍, 彭埃天, 宋晓兵, 凌金锋, 陈霞. 2016. 柑橘黄龙病的传播介体-柑橘木虱在广东果园的发生调查. 植物保护, 42 (1):189-192,196. | |
| [10] |
da Graca J V, Douhan G W, Halbert S E, Keremane M L, Lee R F, Vidalakis G, Zhao H W. 2016. Huanglongbing:an overview of a complex pathosystem ravaging the world’s citrus. Journal of Integrative Plant Biology, 58 (4):373-387.
doi: 10.1111/jipb.12437 URL |
| [11] |
Damsteegt V D, Postnikovae N, Stone A L, Kuhlmann M, Wilson C, Sechler A, Schaad N W, Brlansky R H, Schneider W L. 2010. Murraya paniculata and related species as potential hosts and inoculum reservoirs of‘Candidatus Liberibacter asiaticus’causal agent of Huanglongbing. Plant Disease, 94 (5):528-533.
doi: 10.1094/PDIS-94-5-0528 pmid: 30754478 |
| [12] |
Delessert C, Kazan K, Wilson I W, Straeten D V D, Manners J, Dennis E S, Dolferus R. 2005. The transcription factor ATAF2 represses the expression of pathogenesis-related genes in Arabidopsis. The Plant Journal, 43 (5):745-757.
doi: 10.1111/j.1365-313X.2005.02488.x URL |
| [13] |
Duval M, Hsieh T F, Kim S Y, Thomas T L. 2002. Molecular characterization of AtNAM:a member of the Arabidopsis NAC domain superfamily. Plant Mol Biol, 50 (2):237-248.
doi: 10.1023/A:1016028530943 URL |
| [14] |
Ernst H A, Olsen A N, Skriver K, Larsen S, Leggio L L. 2004. Structure of the conserved domain of ANAC,a member of the NAC family of transcription factors. Embo Reports, 5 (3):297-303.
doi: 10.1038/sj.embor.7400093 URL |
| [15] | Fan Jing. 2007. Molecular cloning and expression pattern of CsCP and CsNAC from‘Navel’orange related to citrus peel pitting[Ph. D. Dissertation]. Chongqing: Chongqing University. (in Chinese) |
| 祝光涛. 2015. 番茄重要农艺性状的全基因组关联分析及野生种质在栽培种的渐渗分析[博士论文]. 重庆: 重庆大学. | |
| [16] | Guo Wen-fang, Liu De-chun, Yang Li, Zhuang Xia, Zhang Juan-juan, Wang Shu-sheng, Liu Yong. 2015. Cloning and expression analysis of new stress-resistant NAC83 gene from citrus. Acta Horticulturae Sinica, 42 (3):445-454. (in Chinese) |
| 郭文芳, 刘德春, 杨莉, 庄霞, 张涓涓, 王书胜, 刘勇. 2015. 柑橘抗逆基因NAC83的克隆与表达分析. 园艺学报, 42 (3):445-454. | |
| [17] |
Halbert S E, Manjunath K L. 2004. Asian citrus psyllids(Sternor-rhyncha:Psyllidae)and greening disease of citrus:a literature review and assessment of risk in Florida. Florida Entomologist, 87 (3):330-353.
doi: 10.1653/0015-4040(2004)087[0330:ACPSPA]2.0.CO;2 URL |
| [18] |
Hu R B, Qi G, Kong Y Z, Kong D J, Gao Q, Zhou G K. 2010. Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. BMC Plant Biology, 10 (1):145.
doi: 10.1186/1471-2229-10-145 URL |
| [19] |
Johnson E G, Wu J, Bright D B, Graham J H. 2014. Association of‘Candidatus Liberibacter asiaticus’root infection,but not phloem plugging with root loss on Huanglongbing-affected trees prior to appearance of foliar symptoms. Plant Pathology, 63 (5):290-298.
doi: 10.1111/ppa.12109 URL |
| [20] |
Kumar S, Stecher G, Tamura K. 2016. MEGA7:molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33 (7):1870-1874.
doi: 10.1093/molbev/msw054 URL |
| [21] |
Le D T, Nishiyama R, Watanabe Y, Mochida K, Yamaguchi S K, Shinozaki K, Tran L S. 2011. Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Research, 18 (4):263-276.
doi: 10.1093/dnares/dsr015 URL |
| [22] |
Lee M H, Jeon H S, Kim H G, Park O K. 2017. An Arabidopsis NAC transcription factor NAC4 promotes pathogen-induced cell death under negative regulation by microRNA164. New Phytologist, 214 (1):343-360.
doi: 10.1111/nph.14371 URL |
| [23] | Li Cheng-hui, Cai Bin. 2013. Genome-wide analysis of NAC transcription factor gene family in grape. Acta Agriculturae Shanghai, 29 (6):7-13. (in Chinese) |
| 李成慧, 蔡斌. 2013. 葡萄NAC转录因子家族全基因组分析. 上海农业学报, 29 (6):7-13. | |
| [24] |
Lin R M, Zhao W S, Meng X B, Wang M, Peng Y L. 2007. Rice gene OsNAC19 encodes a novel NAC-domain transcription factor and responds to infection by Magnaporthe grisea. Plant Science, 172 (1):120-130.
doi: 10.1016/j.plantsci.2006.07.019 URL |
| [25] |
Liu Y Z, Baig M N R, Fan R, Ye J L, Cao Y C, Deng X X. 2009. Identification and expression pattern of a novel NAM,ATAF,and CUC-like gene from Citrus sinensis osbeck. Plant Molecular Biology Reporter, 27 (3):292-297.
doi: 10.1007/s11105-008-0082-z URL |
| [26] | Liu Zhan-ji, Shao Feng-xia, Tang Gui-ying. 2007. The research progress of structure function and regulation of plant NAC transcription factors. Acta Bot Boreal-Occident Sin, 27 (9):1915-1920. (in Chinese) |
| 柳展基, 邵凤霞, 唐桂英. 2007. 植物NAC转录因子的结构功能及其表达调控研究进展. 西北植物学报, 27 (9):1915-1920. | |
| [27] |
Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods, 25 (4):402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [28] |
Nakashima K, Tran L P, Nguyen D V, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamaguchi-Shinozaki K. 2007. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. The Plant Journal, 51 (4):617-630.
doi: 10.1111/j.1365-313X.2007.03168.x URL |
| [29] |
Nuruzzaman M, Manimekalai R, Sharoni A M, Satoh K, Kondoh K, Ooka H, Kikuchi S. 2010. Genome-wide analysis of NAC transcription factor family in rice. Gene, 465 (1):30-44.
doi: 10.1016/j.gene.2010.06.008 URL |
| [30] |
Nuruzzaman M, Sharoni A M, Kikuchi S. 2013. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Frontiers in Microbiology, 4:248.
doi: 10.3389/fmicb.2013.00248 pmid: 24058359 |
| [31] |
Olsen A N, Ernst H A, Leggio L L, Skriver K. 2005. NAC transcription factors:structurally distinct,functionally diverse. Trends Plant Sci, 10 (2):79-87.
doi: 10.1016/j.tplants.2004.12.010 URL |
| [32] |
Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, Hayashizaki Y, Suzuki K, Kojima K, Takahara Y, Yamatomo K, Kikuchi S. 2003. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Research, 10 (6):239-247.
doi: 10.1093/dnares/10.6.239 URL |
| [33] |
Peng A H, Zou X P, He Y R, Chen S C, Liu X F, Zhang J Y, Zhang Q W, Xie Z, Long J H, Zhao X C. 2021. Overexpressing a NPR1-like gene from Citrus paradisi enhanced Huanglongbing resistance in C. sinensis. Plant Cell Reports, 40 (3):529-541.
doi: 10.1007/s00299-020-02648-3 URL |
| [34] | Peng Hui, Yu Xing-wang, Cheng Hui-ying, Zhang Hua, Shi Qing-hua, Li Jian-gui, Ma Hao. 2010. Review on the family of plant NAC transcription factors. Chinese Bulletin of Botany, 45 (2):236-248. (in Chinese) |
| 彭辉, 于兴旺, 成慧颖, 张桦, 石庆华, 李建贵, 麻浩. 2010. 植物NAC转录因子家族研究概况. 植物学报, 45 (2):236-248. | |
| [35] | Qiu Zhi-yu, Huang Hong-lan, Shu Chang, Qiu Jun-jie. 2015. Citrus yellow dragon disease pathogenesis,symptoms and prevention measure. Biological Disaster Science, 38 (3):193-200. (in Chinese) |
| 邱志燏, 黄红兰, 舒畅, 邱俊洁. 2015. 柑橘黄龙病发病机理、症状及防控措施. 生物灾害科学, 38 (3):193-200. | |
| [36] |
Ren T, Qu F, Morris T J. 2000. HRT gene function requires interaction between a NAC protein and viral capsid protein to confer resistance to turnip crinkle virus. Plant Cell, 12 (10):1917-1926.
pmid: 11041886 |
| [37] | Rong Huan, Ren Shi-jie, Wang Zi-ping, Wang Fei, Zhou Yong. 2020. Advances in the structure and function of plant NAC transcription factors. Jiangsu Agricultural Sciences, 48 (18):44-53. (in Chinese) |
| 荣欢, 任师杰, 汪梓坪, 王飞, 周勇. 2020. 植物NAC转录因子的结构及功能研究进展. 江苏农业科学, 48 (18):44-53. | |
| [38] |
Rushton P J, Bokowiec M T, Han S, Zhang H, Brannock J F, Chen X, Laudeman T W, Timko M P. 2008. Tobacco transcription factors:novel insights into transcriptional regulation in the Solanaceae. Plant Physiology, 147 (1):280-295.
doi: 10.1104/pp.107.114041 pmid: 18337489 |
| [39] |
Seo P J, Kim M J, Park J Y, Kim S Y, Jeon J, Lee Y H, Kim J, Park C M. 2010. Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. The Plant Journal, 61 (4):661-671.
doi: 10.1111/j.1365-313X.2009.04091.x URL |
| [40] |
Su H Y, Zhang S Z, Yin Y L, Zhu D Z, Han L Y. 2015. Genome-wide analysis of NAM-ATAF1,2-CUC2 transcription factor family in Solanum lycopersicum. Journal of Plant Biochemistry and Biotechnology, 24 (2):176-183.
doi: 10.1007/s13562-014-0255-9 URL |
| [41] |
Sun H, Hu M L, Li J Y, Chen L, Li M, Zhang S Q, Zhang X L, Yang X Y. 2018. Comprehensive analysis of NAC transcription factors uncovers their roles during fiber development and stress response in cotton. BMC Plant Biology, 18 (1):1-15.
doi: 10.1186/s12870-017-1213-1 URL |
| [42] |
Sun L J, Zhang H J, Li A Y, Huang L, Hong Y B, Ding X S, Nelson R S, Zhou X P, Song F M. 2013. Functions of rice NAC transcriptional factors,ONAC122 and ONAC131,in defense responses against Magnaporthe grisea. Plant Mol Biol, 81 (1):41-56.
doi: 10.1007/s11103-012-9981-3 URL |
| [43] | Sun Li-jun, Li Da-yong, Zhang Hui-juan, Song Feng-ming. 2012. Functions of NAC transcription factors in biotic and abiotic stress responses in plants. Hereditas, 34 (8):993-1002. (in Chinese) |
| 孙利军, 李大勇, 张慧娟, 宋凤鸣. 2012. NAC转录因子在植物抗病和抗非生物胁迫反应中的作用. 遗传, 34 (8):993-1002. | |
| [44] |
Vand S H, Abdullah T L, Sijam k, Abdullah S N, Abdullah N A. 2009. Differential reaction of citrus species in Malaysia to Huanglongbing(HLB) disease using grafting method. American Journal of Agricultural and Biological Sciences, 4 (1):32-38.
doi: 10.3844/ajabssp.2009.32.38 URL |
| [45] |
Wang X E, Basnayake B M, Vindhya S, Zhang H J, Li G J, Li W, Virk N, Mengiste T, Song F M. 2009. The Arabidopsis ATAF1,a NAC transcription factor,is a negative regulator of defense responses against necrotrophic fungal and bacterial pathogens. Molecular Plant-microbe Interactions, 22 (10):1227-1238.
doi: 10.1094/MPMI-22-10-1227 URL |
| [46] |
Wu Y R, Deng Z Y, Lai J B, Zhang Y Y, Yang C P, Yin B J, Zhao Q Z, Zhang L, Li Y, Yang C W, Xie Q. 2009. Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Research, 19 (11):1279-1290.
doi: 10.1038/cr.2009.108 URL |
| [47] |
Xie Z, Zhao K, Long J H, Zhen L, Zou X P, Chen S C. 2021. Comparative analysis of Wanjincheng orange leaf and root responses to‘Candidatus liberibacter asiaticus’infection using leaf-disc grafting. Horticultural Plant Journal, 7 (5):401-410.
doi: 10.1016/j.hpj.2021.01.007 URL |
| [48] | Xie Zhu, Zhao Ke, Zheng Lin, Long Junhong, Du Meixia, He Yongrui, Chen Shanchun, Zou Xiuping. 2020. Cloning and expression analysis of alcohol dehydrogenase CsADH1gene responding to Huanglongbing infection in Citrus. Acta Horticulturae Sinica, 47 (3):445-454. (in Chinese) |
| 谢竹, 赵珂, 郑林, 龙俊宏, 杜美霞, 何永睿, 陈善春, 邹修平. 2020. 响应黄龙病侵染的柑橘乙醇脱氢酶基因CsADH1的克隆与表达分析. 园艺学报, 47 (3):445-454. | |
| [49] |
Xu Q, Chen L L, Ruan X A, Chen D J, Zhu A D, Chen C L, Bertrand D, Jiao W B, Hao B H, Lyon M P, Chen J J, Gao S, Xing F, Lan H, Chang J W, Ge X H, Lei Y, Hu Q, Miao Y, Wang L, Xiao S X, Biswas M K, Zeng W F, Guo F, Cao H B, Yang X M, Xu X W, Cheng Y J, Xu J, Liu J H, Luo J H, Tang Z H, Guo W W, Kuang H H, Zhang H Y, Roose M L, Nagarajan N, Deng X X, Ruan Y J. 2013. The draft genome of sweet orange(Citrus sinensis). Nature Genetics, 45 (1):59-66.
doi: 10.1038/ng.2472 URL |
| [50] |
Yoshii M, Shimizu T, Yamazaki M, Higashi T, Miyao A, Hirochika H, Omura T. 2009. Disruption of a novel gene for a NAC-domain protein in rice confers resistance to rice dwarf virus. The Plant Journal, 57 (4):615-625.
doi: 10.1111/j.1365-313X.2008.03712.x URL |
| [51] |
Yoshii M, Yamazaki M, Rakwal R, KIshi-kaboshi M, Miyaol A, Hirohiko H. 2010. The NAC transcription factor RIM1 of rice is a new regulator of jasmonate signaling. The Plant Journal, 61 (5):804-815.
doi: 10.1111/j.1365-313X.2009.04107.x URL |
| [52] |
Yu C S, Lin C J, Kwang J K. 2004. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Science, 13 (5):1402-1406.
doi: 10.1110/ps.03479604 URL |
| [53] |
Yuan X, Wang H, Cai J T, Li D Y, Song F M. 2019. NAC transcription factors in plant immunity. Phytopathology Research, 1 (1):1-13.
doi: 10.1186/s42483-018-0011-5 URL |
| [54] |
Zou X P, Bai X J, Wen Q L, Xie Z, Wu L, Peng A H, He Y R, Xu L Z, Chen S C. 2019. Comparative analysis of tolerant and susceptible citrus reveals the role of methyl salicylate signaling in the response to Huanglongbing. Journal of Plant Growth Regulation, 38 (4):1516-1528.
doi: 10.1007/s00344-019-09953-6 URL |
| [55] |
Zou X P, Jiang X Y, Xu L Z, Lei T G, Peng A H, He Y R, Yao L X, Chen S C. 2017. Transgenic citrus expressing synthesized cecropin B genes in the phloem exhibits decreased susceptibility to Huanglongbing. Plant Molecular Biology, 93 (4):341-353.
doi: 10.1007/s11103-016-0565-5 URL |
| [56] |
Zou X P, Zhao K, Liu Y N, Du M X, Zheng L, Wang S, Xu L Z, Peng A H, He Y R, Long Q, Chen S C. 2021. Overexpression of salicylic acid carboxyl methyltransferase(CsSAMT1)enhances tolerance to Huanglongbing disease in Wanjincheng orange[Citrus sinensis(L.)Osbeck]. International Journal of Molecular Sciences, 22 (6):2803.
doi: 10.3390/ijms22062803 URL |
| [1] | ZHAO Xueyan, WANG Qi, WANG Li, WANG Fangyuan, WANG Qing, LI Yan. Comparative Transcriptome Analysis of Differential Expression in Different Tissues of Corydalis yanhusuo [J]. Acta Horticulturae Sinica, 2023, 50(1): 177-187. |
| [2] | LI Zhenxi, PAN Ruixuan, XU Meirong, ZHENG Zheng, DENG Xiaoling. Development of Duplex Real-time PCR Assay of‘Candidatus Liberibacter asiatics’ [J]. Acta Horticulturae Sinica, 2023, 50(1): 188-196. |
| [3] | LIU Zhiyuan, XU Zhaosheng, ZHANG Helong, and QIAN Wei. A New Oversummer Spinach Cultivar‘Shubo 16’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 95-96. |
| [4] | WANG Sha, ZHANG Xinhui, ZHAO Yujie, LI Bianbian, ZHAO Xueqing, SHEN Yu, DONG Jianmei, YUAN Zhaohe. Cloning and Functional Analysis of PgMYB111 Related to Anthocyanin Synthesis in Pomegranate [J]. Acta Horticulturae Sinica, 2022, 49(9): 1883-1894. |
| [5] | GAO Yanlong, WU Yuxia, ZHANG Zhongxing, WANG Shuangcheng, ZHANG Rui, ZHANG De, WANG Yanxiu. Bioinformatics Analysis of Apple ELO Gene Family and Its Expression Analysis Under Low Temperature Stress [J]. Acta Horticulturae Sinica, 2022, 49(8): 1621-1636. |
| [6] | QIU Ziwen, LIU Linmin, LIN Yongsheng, LIN Xiaojie, LI Yongyu, WU Shaohua, YANG Chao. Cloning and Functional Analysis of the MbEGS Gene from Melaleuca bracteata [J]. Acta Horticulturae Sinica, 2022, 49(8): 1747-1760. |
| [7] | MA Weifeng, LI Yanmei, MA Zonghuan, CHEN Baihong, MAO Juan. Identification of Apple POD Gene Family and Functional Analysis of MdPOD15 Gene [J]. Acta Horticulturae Sinica, 2022, 49(6): 1181-1199. |
| [8] | ZHANG Kai, MA Mingying, WANG Ping, LI Yi, JIN Yan, SHENG Ling, DENG Ziniu, MA Xianfeng. Identification of HSP20 Family Genes in Citrus and Their Expression in Pathogen Infection Responses Citrus Canker [J]. Acta Horticulturae Sinica, 2022, 49(6): 1213-1232. |
| [9] | LIU Yaoyao, WU Yanyan, Shi Yan, MAO Tianyu, BAO Manzhu, ZHANG Junwei, ZHANG Jie. Preliminary Study on the Relationship Between Promoter Sequence Difference of PmTAC1 and Weeping Trait of Prunus mume [J]. Acta Horticulturae Sinica, 2022, 49(6): 1327-1338. |
| [10] | LIANG Chen, SUN Ruyi, XIANG Rui, SUN Yimeng, SHI Xiaoxin, DU Guoqiang, WANG Li. Genome-wide Identification of Grape GRF Family and Expression Analysis [J]. Acta Horticulturae Sinica, 2022, 49(5): 995-1007. |
| [11] | XIAO Xuechen, LIU Mengyu, JIANG Mengqi, CHEN Yan, XUE Xiaodong, ZHOU Chengzhe, WU Xingjian, WU Junnan, GUO Yinsheng, YEH Kaiwen, LAI Zhongxiong, LIN Yuling. Whole-genome Identification and Expression Analysis of SNAT,ASMT and COMT Families of Melatonin Synthesis Pathway in Dimocarpus longan [J]. Acta Horticulturae Sinica, 2022, 49(5): 1031-1046. |
| [12] | WEI Zhuangmin, WEI Sijia, CHEN Peng, HU Jianbing, TANG Yuqing, YE Junli, LI Xianxin, DENG Xiuxin, CHAI Lijun. Identification of S-genotypes of 63 Pummelo Germplasm Resources [J]. Acta Horticulturae Sinica, 2022, 49(5): 1111-1120. |
| [13] | GAO Weilin, ZHANG Liman, XUE Chaoling, ZHANG Yao, LIU Mengjun, ZHAO Jin. Expression of E-type MADS-box Genes in Flower and Fruits and Protein Interaction Analysis in Chinese Jujube [J]. Acta Horticulturae Sinica, 2022, 49(4): 739-748. |
| [14] | WANG Ying, QIN Yangyang, ZENG Ting, LIAO Ping, ZHANG Wei, ZHOU Yan, ZHOU Changyong. Effects of Citrus Yellow Vein Clearing Virus on Photosynthetic Characteristics and Chloroplast Ultrastructure of Lemon [J]. Acta Horticulturae Sinica, 2022, 49(4): 861-867. |
| [15] | LIU Mengyu, JIANG Mengqi, CHEN Yan, ZHANG Shuting, XUE Xiaodong, XIAO Xuechen, LAI Zhongxiong, LIN Yuling. Genome-wide Identification and Expression Analysis of GDSL Esterase/Lipase Genes in Dimocarpus longan [J]. Acta Horticulturae Sinica, 2022, 49(3): 597-612. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd