
Acta Horticulturae Sinica ›› 2022, Vol. 49 ›› Issue (6): 1301-1312.doi: 10.16420/j.issn.0513-353x.2021-0629
• Research Papers • Previous Articles Next Articles
CHEN Daozong, LIU Yi, SHEN Wenjie, ZHU Bo, TAN Chen( )
)
Received:2021-06-29
Revised:2021-11-27
Online:2022-06-25
Published:2022-07-05
Contact:
TAN Chen
E-mail:tanchen2020@gnnu.edu.cn
CLC Number:
CHEN Daozong, LIU Yi, SHEN Wenjie, ZHU Bo, TAN Chen. Identification and Analysis of PAP1/2 Homologous Genes in Brassica rapa,B. oleracea and B. napus[J]. Acta Horticulturae Sinica, 2022, 49(6): 1301-1312.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2021-0629
| Function | 拟南芥 A. thaliana | 白菜 B. rapa | 甘蓝 B. oleracea | 甘蓝型油菜-AA B. napus-AA | 甘蓝型油菜-CC B. napus-CC |
|---|---|---|---|---|---|
| PAP1(MYB75) | AT1G56650 | BraA02g017040.3C | Bo00835s060.1 | BnaA02G0160600ZS | BnaC02G0205500ZS |
| PAP1(MYB75) | AT1G56650 | BraA03g040840.3C | Bo3g081880.1 | BnaA03G0376600ZS | BnaC03G0461400ZS |
| PAP1(MYB75) | AT1G56650 | Bo6g099880.1 | BnaA07G0286900ZS | BnaC06G0328400ZS | |
| PAP1(MYB75) | AT1G56650 | BnaC06G0328700ZS | |||
| PAP2(MYB90) | AT1G66390 | BraA07g032100.3C | Bo6g100940.1 | BnaA07G0287000ZS | BnaC06G0329100ZS |
Table 1 Copy number and collinearity of PAP1/2 homologous genes in Arabidopsis,Brassica rapa,B. oleracea and B. napus
| Function | 拟南芥 A. thaliana | 白菜 B. rapa | 甘蓝 B. oleracea | 甘蓝型油菜-AA B. napus-AA | 甘蓝型油菜-CC B. napus-CC |
|---|---|---|---|---|---|
| PAP1(MYB75) | AT1G56650 | BraA02g017040.3C | Bo00835s060.1 | BnaA02G0160600ZS | BnaC02G0205500ZS |
| PAP1(MYB75) | AT1G56650 | BraA03g040840.3C | Bo3g081880.1 | BnaA03G0376600ZS | BnaC03G0461400ZS |
| PAP1(MYB75) | AT1G56650 | Bo6g099880.1 | BnaA07G0286900ZS | BnaC06G0328400ZS | |
| PAP1(MYB75) | AT1G56650 | BnaC06G0328700ZS | |||
| PAP2(MYB90) | AT1G66390 | BraA07g032100.3C | Bo6g100940.1 | BnaA07G0287000ZS | BnaC06G0329100ZS |
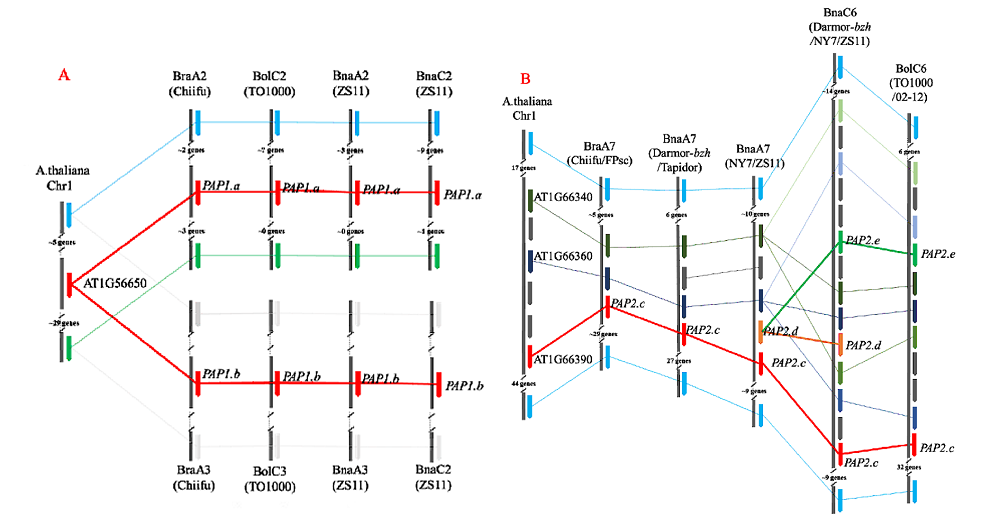
Fig. 1 The comparison map of the segment of PAP1/2 homologous genes in Arabidopsis,Brassica rapa,B. oleracea and B. napus A. The comparison map of the segment of PAP1/2 homologous genes on A2-C2,A3-C3 chromosome;B. The comparison map of the segment of PAP1/2 homologous genes on A2-C6 chromosome.
| 拟南芥 A. thaliana | 白菜B. rapa Chiifu-A2 | 甘蓝B. oleracea TO1000-C2 | 甘蓝型油菜B. napus ZS11-A2 | 甘蓝型油菜B. napus ZS11-C2 | |
|---|---|---|---|---|---|
| A2-C2 | AT1G66330 | BraA02g017010.3C | Bo2g052820.1 | BnaA02G0160200ZS | BnaC02T0204500ZS |
| AT1G66390 | BraA02g017040.3C | Bo00835s060.1 | BnaA02T0160600ZS | BnaC02T0205500ZS | |
| AT1G66470 | BraA02g017080.3C | Bo00835s050.1 | BnaA02T0160700ZS | BnaC02T0205700ZS | |
| A3-C3 | AT1G66330 | Null | Null | Null | Null |
| AT1G56650 | BraA03g040840.3C | Bo3g081880.1 | BnaA03G0376600ZS | BnaC03G0461400ZS | |
| AT1G66470 | Null | Null | Null | Null | |
| A7-C6 | AT1G66210 | BraA07g032000.3C | Bo6g099780.1 | BnaA07T0285500ZS | BnaC06T0327300ZS |
| AT1G66390 | BraA07g032100.3C | Bo6g099880.1 | BnaA07G0286900ZS | BnaC06G0328400ZS | |
| AT1G66390 | BraA07g032100.3C | Bo6g099880.1 | BnaA07G0286900ZS | BnaC06G0328700ZS | |
| AT1G66390 | BraA07g032100.3C | Bo6g099880.1 | BnaA07G0287000ZS | BnaC06G0329100ZS | |
| AT1G66840 | BraA07g032410.3C | Bo6g101320.1 | BnaA07T0289800ZS | BnaC06T0333000ZS |
Table 2 The comparison map of the segment of PAP1/2 homologous genes in Arabidopsis,Brassica rapa,B. oleracea and B. napus
| 拟南芥 A. thaliana | 白菜B. rapa Chiifu-A2 | 甘蓝B. oleracea TO1000-C2 | 甘蓝型油菜B. napus ZS11-A2 | 甘蓝型油菜B. napus ZS11-C2 | |
|---|---|---|---|---|---|
| A2-C2 | AT1G66330 | BraA02g017010.3C | Bo2g052820.1 | BnaA02G0160200ZS | BnaC02T0204500ZS |
| AT1G66390 | BraA02g017040.3C | Bo00835s060.1 | BnaA02T0160600ZS | BnaC02T0205500ZS | |
| AT1G66470 | BraA02g017080.3C | Bo00835s050.1 | BnaA02T0160700ZS | BnaC02T0205700ZS | |
| A3-C3 | AT1G66330 | Null | Null | Null | Null |
| AT1G56650 | BraA03g040840.3C | Bo3g081880.1 | BnaA03G0376600ZS | BnaC03G0461400ZS | |
| AT1G66470 | Null | Null | Null | Null | |
| A7-C6 | AT1G66210 | BraA07g032000.3C | Bo6g099780.1 | BnaA07T0285500ZS | BnaC06T0327300ZS |
| AT1G66390 | BraA07g032100.3C | Bo6g099880.1 | BnaA07G0286900ZS | BnaC06G0328400ZS | |
| AT1G66390 | BraA07g032100.3C | Bo6g099880.1 | BnaA07G0286900ZS | BnaC06G0328700ZS | |
| AT1G66390 | BraA07g032100.3C | Bo6g099880.1 | BnaA07G0287000ZS | BnaC06G0329100ZS | |
| AT1G66840 | BraA07g032410.3C | Bo6g101320.1 | BnaA07T0289800ZS | BnaC06T0333000ZS |
| 保守结构域名称 Motif name | 保守结构域序列 Motif sequence | e值 e-value | 宽度 Width |
|---|---|---|---|
| Motif 1 | C[KN]T[KQ]M[KR]KRN[IV][PT][CF]S[SY]T[TQ]PAQK[IN][DE]V[FL]KPRPR[SL] FTVNNGCSH[LIF][NH]G[ML]P[EK][VA]D[VI][VI]P | 1.4e-672 | 50 |
| Motif 2 | [YS]ELV[SN]NLMDG[EQ]N[MR]WW[EK]SLL[ED]ES[QK][DEQ]P[AD][AG][LI][VF] P[EK][AGS]T[AE]T[KE]K[GL]AT[SF]AFDVEQLW[SN] | 3.1e-601 | 50 |
| Motif 3 | AWTAEED[SN]LLR[QR]CIDKYGEGKWHQVPLRAGL | 8.7e-512 | 31 |
| Motif 4 | K[LF][SN]SDEVDLLLRLHKLLGNRWSLIAGRLPGRTAND[VI]KNYWNTHLSKKHEP | 7.6e-799 | 50 |
| Motif 5 | RCRKSCRLRWLNYLKPSI[KN][RK]G | 3.0e-340 | 21 |
| Motif 6 | [ND][TIA]NNV[CS]EN[SI][IFM]T[CY][NK][KN]D | 2.0e-156 | 15 |
| Motif 7 | ME[GD]S[SP]KGL[RKT]KG | 1.2e-113 | 11 |
| Motif 8 | [LM]LDGET[VG][ET] | 7.2e-62 | 8 |
| Motif 9 | [LP][CF]LG[LFV]N | 1.4e-20 | 6 |
Table 3 Conserved motif sequence of PAP1/2 homologous genes in Arabidopsis,Brassica rapa,B. oleracea and B. napus
| 保守结构域名称 Motif name | 保守结构域序列 Motif sequence | e值 e-value | 宽度 Width |
|---|---|---|---|
| Motif 1 | C[KN]T[KQ]M[KR]KRN[IV][PT][CF]S[SY]T[TQ]PAQK[IN][DE]V[FL]KPRPR[SL] FTVNNGCSH[LIF][NH]G[ML]P[EK][VA]D[VI][VI]P | 1.4e-672 | 50 |
| Motif 2 | [YS]ELV[SN]NLMDG[EQ]N[MR]WW[EK]SLL[ED]ES[QK][DEQ]P[AD][AG][LI][VF] P[EK][AGS]T[AE]T[KE]K[GL]AT[SF]AFDVEQLW[SN] | 3.1e-601 | 50 |
| Motif 3 | AWTAEED[SN]LLR[QR]CIDKYGEGKWHQVPLRAGL | 8.7e-512 | 31 |
| Motif 4 | K[LF][SN]SDEVDLLLRLHKLLGNRWSLIAGRLPGRTAND[VI]KNYWNTHLSKKHEP | 7.6e-799 | 50 |
| Motif 5 | RCRKSCRLRWLNYLKPSI[KN][RK]G | 3.0e-340 | 21 |
| Motif 6 | [ND][TIA]NNV[CS]EN[SI][IFM]T[CY][NK][KN]D | 2.0e-156 | 15 |
| Motif 7 | ME[GD]S[SP]KGL[RKT]KG | 1.2e-113 | 11 |
| Motif 8 | [LM]LDGET[VG][ET] | 7.2e-62 | 8 |
| Motif 9 | [LP][CF]LG[LFV]N | 1.4e-20 | 6 |
| [1] |
Bolger A M, Lohse M, Usadel B. 2014. Trimmomatic:a flexible trimmer for illumina sequence data. Bioinformatics, 30 (15):2114-2120.
doi: 10.1093/bioinformatics/btu170 URL |
| [2] |
Borevitz J O, Xia Y, Blount J, Dixon R A, Lamb C. 2000. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell, 12 (12):2383-2394.
pmid: 11148285 |
| [3] |
Chalhoub B, Denoeud F, Liu S, Parkin I A, Tang H, Wang X, Chiquet J, Belcram H, Tong C, Samans B, Corréa M, Da Silva C, Just J, Falentin C, Koh C S, Le Clainche I, Bernard M, Bento P, Noel B, Labadie K, Alberti A, Charles M, Arnaud D, Guo H, Daviaud C, Alamery S, Jabbari K, Zhao M, Edger P P, Chelaifa H, Tack D, Lassalle G, Mestiri I, Schnel N, Le Paslier M C, Fan G, Renault V, Bayer P E, Golicz A A, Manoli S, Lee T H, Thi V H, Chalabi S, Hu Q, Fan C, Tollenaere R, Lu Y, Battail C, Shen J, Sidebottom C H, Wang X, Canaguier A, Chauveau A, Bérard A, Deniot G, Guan M, Liu Z, Sun F, Lim Y P, Lyons E, Town C D, Bancroft I, Wang X, Meng J, Ma J, Pires J C, King G J, Brunel D, Delourme R, Renard M, Aury J M, Adams K L, Batley J, Snowdon R J, Tost J, Edwards D, Zhou Y, Hua W, Sharpe A G, Paterson A H, Guan C, Wincker P. 2014. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science, 345 (6199):950-953.
doi: 10.1126/science.1253435 pmid: 25146293 |
| [4] |
Chen D Z, Liu Y, Yin S, Qiu J, Jin Q D, King G J, Wang J, Ge X H, Li Z Y. 2020. Identification and functional analysis of BnaPAP2.A7 revealed that its alternative splicing isoforms play opposite roles in anthocyanin biosynthesis in Brassica napus L. Front Plant Sci, 11:983.
doi: 10.3389/fpls.2020.00983 URL |
| [5] | Cheng F, Wu J, Fang L, Wang X. 2012. Syntenic gene analysis between Brassica rapa and other Brassicaceae species. Front Plant Sci, 30:198. |
| [6] | Cheng Y, Zhu W, Chen Y, Ito S, Asami T, Wang X. 2014. Brassinosteroids control root epidermal cell fate via direct regulation of a MYB-bHLH-WD 40 complex by GSK3-like kinases. Elife, 25:e02525. |
| [7] |
Cone K C, Burr F A, Burr B. 1986. Molecular analysis of the maize anthocyanin regulatory locus C1. Proc Natl Acad Sci USA, 83:9631-9635.
doi: 10.1073/pnas.83.24.9631 URL |
| [8] |
Cone K C, Cocciolone S M, Burr F A, Burr B. 1993. Maize anthocyanin regulatory gene pl is a duplicate of c 1 that functions in the plant. Plant Cell, 5 (12):1795-1805.
pmid: 8305872 |
| [9] |
Devic M, Guilleminot J, Debeaujon I, Bechtold N, Bensaude E, Koornneef M, Pelletier G, Delseny M. 1999. The BANYULS gene encodes a DFR-like protein and is a marker of early seed coat development. Plant J, 19 (4):387-398.
pmid: 10504561 |
| [10] |
Fu Z Z, Shang H Q, Jiang H, Gao J, Dong X Y, Wang H J, Li Y M, Wang L M, Zhang J, Shu Q Y, Chao Y C, Xu M L, Wang R, Wang L S, Zhang H C. 2020. Systematic identification of the light-quality responding anthocyanin synthesis-related transcripts in petunia petals. Horticultural Plant Journal, 6 (6):428-438.
doi: 10.1016/j.hpj.2020.11.006 URL |
| [11] |
Guo N, Cheng F, Wu J, Liu B, Zheng S, Liang J, Wang X. 2014. Anthocyanin biosynthetic genes in Brassica rapa. BMC Genomics, 15:426.
doi: 10.1186/1471-2164-15-426 URL |
| [12] |
He Q, Wu J, Xue Y, Zhao W, Li R, Zhang L. 2020. The novel gene BrMYB2,located on chromosome A07,with a short intron 1 controls the purple-head trait of Chinese cabbage(Brassica rapa L.). Hortic Res, 7:97.
doi: 10.1038/s41438-020-0319-z URL |
| [13] |
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg S L. 2013. TopHat2:accurate alignment of transcriptomes in the presence of insertions,deletions and gene fusions. Genome Biol, 14 (4):R36.
doi: 10.1186/gb-2013-14-4-r36 URL |
| [14] |
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X:molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol, 35 (6):1547-1549.
doi: 10.1093/molbev/msy096 URL |
| [15] | Li Chonghui, Yang Guangsui, Zhang Zhiqun, Yin Junmei. 2021. A novel R2R3-MYB transcription factor gene AaMYB6 involved in anthocyanin biosynthesis in Anthurium andraeanum. Acta Horticulturae Sinica, 48 (10):1859-1872. (in Chinese) |
| 李崇晖, 杨光穗, 张志群, 尹俊梅. 2021. 红掌R2R3-MYB转录因子基因AaMYB6调控花青素苷合成. 园艺学报, 48 (10):1859-1872. | |
| [16] |
Li Y M, Zeng X Z, Guo H, Luo Y W, Kear P, Zhang S M, Zhu G T. 2021. Genome-wide analysis of MYB gene family in potato provides insights into tissue-specific regulation of anthocyanin biosynthesis. Horticultural Plant Journal, 7 (2):129-141.
doi: 10.1016/j.hpj.2020.12.001 URL |
| [17] |
Liu S, Liu Y, Yang X, Tong C, Edwards D, Parkin I A, Zhao M, Ma J, Yu J, Huang S, Wang X, Wang J, Lu K, Fang Z, Bancroft I, Yang T J, Hu Q, Wang X, Yue Z, Li H, Yang L, Wu J, Zhou Q, Wang W, King G J, Pires J C, Lu C, Wu Z, Sampath P, Wang Z, Guo H, Pan S, Yang L, Min J, Zhang D, Jin D, Li W, Belcram H, Tu J, Guan M, Qi C, Du D, Li J, Jiang L, Batley J, Sharpe A G, Park B S, Ruperao P, Cheng F, Waminal N E, Huang Y, Dong C, Wang L, Li J, Hu Z, Zhuang M, Huang Y, Huang J, Shi J, Mei D, Liu J, Lee T H, Wang J, Jin H, Li Z, Li X, Zhang J, Xiao L, Zhou Y, Liu Z, Liu X, Qin R, Tang X, Liu W, Wang Y, Zhang Y, Lee J, Kim H H, Denoeud F, Xu X, Liang X, Hua W, Wang X, Wang J, Chalhoub B, Paterson A H. 2014. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat Commun, 5:3930.
doi: 10.1038/ncomms4930 URL |
| [18] | Liu Xin, Chen Yunzhu, Kim Pyol, Kim Min-Jun, Song Hyondok, Li Yuhua, Wang Yu. 2020. Progress on molecular mechanism and regulation of tomato fruit color formation. Acta Horticulturae Sinica, 47 (9):1689-1704. (in Chinese) |
| 刘昕, 陈韵竹, Kim Pyol, Kim Min-Jun, Song Hyondok, 李玉花, 王宇. 2020. 番茄果实颜色形成的分子机制及调控研究进展. 园艺学报, 47 (9):1689-1704. | |
| [19] |
Meng J X, Gao Y, Han M L, Liu P Y, Yang C, Shen T, Li H H. 2020. In vitro anthocyanin induction and metabolite analysis in Malus spectabilis leaves under low nitrogen conditions. Horticultural Plant Journal, 6 (5):284-292.
doi: 10.1016/j.hpj.2020.06.004 URL |
| [20] |
Pertea M, Kim D, Pertea G M, Leek J T, Salzberg S L. 2016. Transcript-level expression analysis of RNA-seq experiments with HISAT,StringTie and Ballgown. Nat Protoc, 11 (9):1650-1667.
doi: 10.1038/nprot.2016.095 URL |
| [21] |
Shi M Z, Xie D Y. 2014. Biosynthesis and metabolic engineering of anthocyanins in Arabidopsis thaliana. Recent Pat Biotechnol, 8 (1):47-60.
doi: 10.2174/1872208307666131218123538 URL |
| [22] |
Tanaka M, Fujimori T, Uchida I, Yamaguchi S, Takeda K. 2001. A malonylated,anthocyanin and flavonols in blue Meconopsis flowers. Phytochemistry, 56 (4):373-386.
pmid: 11249104 |
| [23] |
Tanaka Y, Brugliera F, Chandler S. 2009. Recent progress of flower colour modification by biotechnology. Int J Mol Sci, 10 (12):5350-5369.
doi: 10.3390/ijms10125350 URL |
| [24] |
Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun J H, Bancroft I, Cheng F, Huang S, Li X, Hua W, Wang J, Wang X, Freeling M, Pires J C, Paterson A H, Chalhoub B, Wang B, Hayward A, Sharpe A G, Park B S, Weisshaar B, Liu B, Li B, Liu B, Tong C, Song C, Duran C, Peng C, Geng C, Koh C, Lin C, Edwards D, Mu D, Shen D, Soumpourou E, Li F, Fraser F, Conant G, Lassalle G, King G J, Bonnema G, Tang H, Wang H, Belcram H, Zhou H, Hirakawa H, Abe H, Guo H, Wang H, Jin H, Parkin I A, Batley J, Kim J S, Just J, Li J, Xu J, Deng J, Kim J A, Li J, Yu J, Meng J, Wang J, Min J, Poulain J, Wang J, Hatakeyama K, Wu K, Wang L, Fang L, Trick M, Links M G, Zhao M, Jin M, Ramchiary N, Drou N, Berkman P J, Cai Q, Huang Q, Li R, Tabata S, Cheng S, Zhang S, Zhang S, Huang S, Sato S, Sun S, Kwon S J, Choi S R, Lee T H, Fan W, Zhao X, Tan X, Xu X, Wang Y, Qiu Y, Yin Y, Li Y, Du Y, Liao Y, Lim Y, Narusaka Y, Wang Y, Wang Z, Li Z, Wang Z, Xiong Z, Zhang Z. 2011. Brassica rapa genome sequencing project consortium. The genome of the mesopolyploid crop species Brassica rapa. Nat Genet, 43 (10):1035-1039.
doi: 10.1038/ng.919 URL |
| [25] |
Xi H C, He Y J, Chen H Y. 2021. Functional characterization of SmbHLH13in anthocyanin biosynthesis and flowering in eggplant. Horticultural Plant Journal, 7 (1):73-80.
doi: 10.1016/j.hpj.2020.08.006 URL |
| [26] |
Xu W, Dubos C, Lepiniec L. 2015. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci, 20 (3):176-185.
doi: 10.1016/j.tplants.2014.12.001 URL |
| [27] |
Xu W, Grain D, Bobet S, Le Gourrierec J, Thévenin J, Kelemen Z, Lepiniec L, Dubos C. 2014. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. New Phytol, 202 (1):132-144.
doi: 10.1111/nph.12620 URL |
| [28] |
Yan C, An G, Zhu T, Zhang W, Zhang L, Peng L, Chen J, Kuang H. 2019. Independent activation of the BoMYB2 gene leading to purple traits in Brassica oleracea. Theor Appl Genet, 132 (4):895-906.
doi: 10.1007/s00122-018-3245-9 URL |
| [29] |
Yang J, Liu D, Wang X, Ji C, Cheng F, Liu B, Hu Z, Chen S, Pental D, Ju Y, Yao P, Li X, Xie K, Zhang J, Wang J, Liu F, Ma W, Shopan J, Zheng H, Mackenzie S A, Zhang M. 2016. The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat Genet, 48 (10):1225-1232.
doi: 10.1038/ng.3657 URL |
| [30] |
Zhao L, Gao L, Wang H, Chen X, Wang Y, Yang H, Wei C, Wan X, Xia T. 2013. The R2R3-MYB,bHLH,WD40,and related transcription factors in flavonoid biosynthesis. Funct Integr Genomics, 13 (1):75-98.
doi: 10.1007/s10142-012-0301-4 URL |
| [31] |
Zhou L L, Shi M Z, Xie D Y. 2012. Regulation of anthocyanin biosynthesis by nitrogen in TTG1-GL3/TT8-PAP1-programmed red cells of Arabidopsis thaliana. Planta, 236 (3):825-837.
doi: 10.1007/s00425-012-1674-2 URL |
| [1] | YE Zimao, SHEN Wanxia, LIU Mengyu, WANG Tong, ZHANG Xiaonan, YU Xin, LIU Xiaofeng, and ZHAO Xiaochun, . Effect of R2R3-MYB Transcription Factor CitMYB21 on Flavonoids Biosynthesis in Citrus [J]. Acta Horticulturae Sinica, 2023, 50(2): 250-264. |
| [2] | ZOU Xue, DING Fan, LIU Lifang, YU Hankaizong, CHEN Nianwei, and RAO Liping. A New Purple Potato Cultivar‘Mianziyu 1’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 93-94. |
| [3] | WANG Sha, ZHANG Xinhui, ZHAO Yujie, LI Bianbian, ZHAO Xueqing, SHEN Yu, DONG Jianmei, YUAN Zhaohe. Cloning and Functional Analysis of PgMYB111 Related to Anthocyanin Synthesis in Pomegranate [J]. Acta Horticulturae Sinica, 2022, 49(9): 1883-1894. |
| [4] | HUANG Ling, HU Xianmei, LIANG Zehui, WANG Yanping, CHAN Zhulong, XIANG Lin. Cloning and Function Identification of Anthocyanidin Synthase Gene TgANS in Tulipa gesneriana [J]. Acta Horticulturae Sinica, 2022, 49(9): 1935-1944. |
| [5] | LI Maofu, YANG Yuan, WANG Hua, FAN Youwei, SUN Pei, JIN Wanmei. Analysis the Function of R2R3 MYB Transcription Factor RhMYB113c on Regulating Anthocyanin Synthesis in Rosa hybrida [J]. Acta Horticulturae Sinica, 2022, 49(9): 1957-1966. |
| [6] | QIAN Jieyu, JIANG Lingli, ZHENG Gang, CHEN Jiahong, LAI Wuhao, XU Menghan, FU Jianxin, ZHANG Chao. Identification and Expression Analysis of MYB Transcription Factors Regulating the Anthocyanin Biosynthesis in Zinnia elegans and Function Research of ZeMYB9 [J]. Acta Horticulturae Sinica, 2022, 49(7): 1505-1518. |
| [7] | ZHANG Lugang, LU Qianqian, HE Qiong, XUE Yihua, MA Xiaomin, MA Shuai, NIE Shanshan, YANG Wenjing. Creation of Novel Germplasm of Purple-orange Heading Chinese Cabbage [J]. Acta Horticulturae Sinica, 2022, 49(7): 1582-1588. |
| [8] | LI Xiaoming, YU Junchi, WANG Chunxia. Comparison of Growth and Secondary Metabolites of Purple and White Flower Dracocephalum moldavica Under Field,Greenhouse and Greenhouse Shading Conditions [J]. Acta Horticulturae Sinica, 2022, 49(6): 1363-1370. |
| [9] | LI Lixian, WANG Shuo, CHEN Ying, WU Yingtao, WANG Yaqian, FANG Yue, CHEN Xuesen, TIAN Changping, FENG Shouqian. PavMYB10.1 Promotes Anthocyanin Accumulation by Positively Regulating PavRiant in Sweet Cherry [J]. Acta Horticulturae Sinica, 2022, 49(5): 1023-1030. |
| [10] | DENG Jiao, SU Mengyue, LIU Xuelian, OU Kefang, HU Zhengrong, YANG Pingfang. Transcriptome Analysis Revealed the Formation Mechanism of Floral Color of Lotus‘Dasajin’with Bicolor Petal [J]. Acta Horticulturae Sinica, 2022, 49(2): 365-377. |
| [11] | WANG Zhiyu, CHANG Beibei, LIU Qi, CHENG Xiaofan, DU Xiaoyun, YU Xiaoli, SONG Laiqing, ZHAO Lingling. Study on Expression and Anthocyanin Accumulation of Solute Carrier Gene MdSLC35F2-like in Apple [J]. Acta Horticulturae Sinica, 2022, 49(11): 2293-2303. |
| [12] | SUN Wei, SUN Shiyu, CHEN Yiran, WANG Yuhan, ZHANG Yan, JU Zhigang, YI Yin. Cloning and Function Analysis of Chalcone Isomerase Gene RdCHI1 in Rhododendron delavayi [J]. Acta Horticulturae Sinica, 2022, 49(11): 2407-2418. |
| [13] | HONG Yanhong, YE Qinghua, LI Zekun, WANG Wei, XIE Qian, CHEN Qingxi, CHEN Jianqing. Accumulation of Anthocyanins in Red-flowered Strawberry‘Meihong’Petals and Expression Analysis of MYB Gene [J]. Acta Horticulturae Sinica, 2021, 48(8): 1470-1484. |
| [14] | DING Yunhua, Budahn Holger, ZHAO Hong, ZHAO Xiuyun. Accurate Identification of Radish Chromosome F in the Backcross Progeny of Brassica rapa and Rape-radish Chromosome F Addition Line [J]. Acta Horticulturae Sinica, 2021, 48(7): 1295-1303. |
| [15] | ZHANG Shuangshuang, SU Wei, LIU Yang, WANG Haiping, SONG Jiangping, YANG Wenlong, JIA Huixia, Zhang Xiaohui, LI Xixiang. Germplasm Innovation and Black Rot Resistance Transferring in Brassica rapa ssp. chinensis Through Interspecific Hybridization with Brassica carinata [J]. Acta Horticulturae Sinica, 2021, 48(7): 1304-1316. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd