
Acta Horticulturae Sinica ›› 2022, Vol. 49 ›› Issue (4): 801-815.doi: 10.16420/j.issn.0513-353x.2021-0660
• Research Papers • Previous Articles Next Articles
SHEN Nan, ZHANG Jingcheng, WANG Chengchen, BIAN Yinbing, XIAO Yang*( )
)
Received:2021-08-26
Revised:2022-03-21
Online:2022-04-25
Published:2022-04-24
Contact:
XIAO Yang
E-mail:xiaoyang@mail.hzau.edu.cn
CLC Number:
SHEN Nan, ZHANG Jingcheng, WANG Chengchen, BIAN Yinbing, XIAO Yang. Studies on Transcriptome During Fruiting Body Development of Lentinula edodes[J]. Acta Horticulturae Sinica, 2022, 49(4): 801-815.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2021-0660

Fig. 1 Samples from different development stages of Lentinula edodes for transcriptome sequencing MY:Mycelium with brown film;PR:Primordium;D1PI:Pileus of young fruiting body;D1ST:Stipe of young fruiting body;D2PI:Pileus of mature fruiting body before pileus opening;D2ST:Stipe of mature fruiting body before pileus opening;MFPI:Pileus of mature fruiting body after pileus opening;MFST:Stipe of mature fruiting body after pileus opening. The same below.
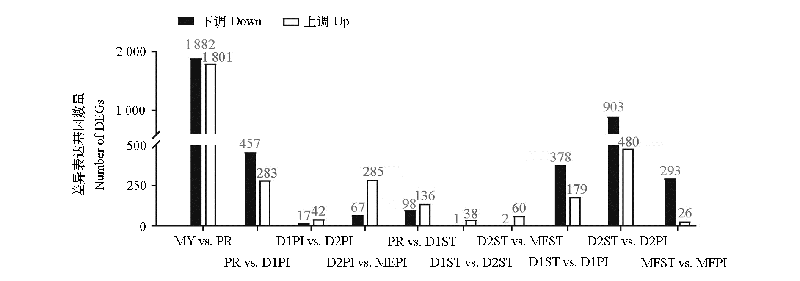
Fig. 2 The number of DEGs between two adjacent developmental stages or tissues(pileus and stipe)at the same stage The code names of samples are consistent with those in Fig. 1.

Fig. 3 The enriched GO terms and heatmap of gene expression of genes in significant profiles In the profile box,the number in the upper left corner represents the profile number and the number in the bottom left corner represents the number of genes in it. The code names of samples are consistent with those in Fig. 1.

Fig. 4 The relationships between gene co-expression modules and different development stages or tissues in Lentinula edodes The code names of samples are consistent with those in Fig. 1.
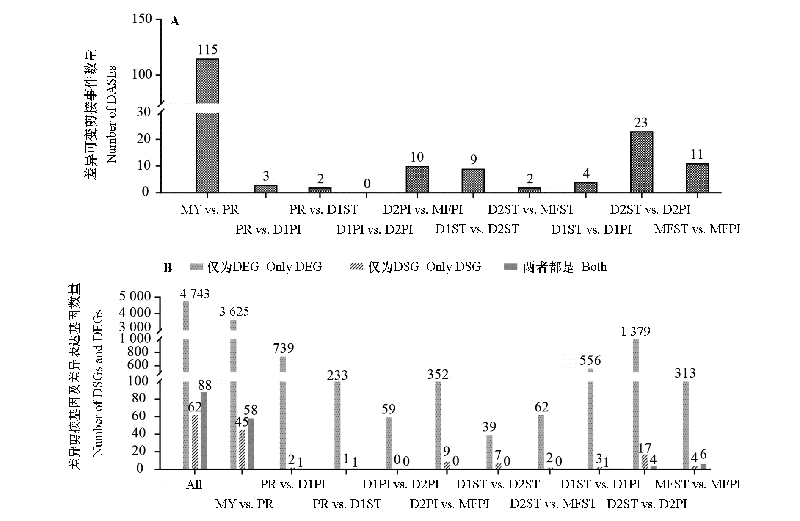
Fig. 5 The number of DASEs(A),DSGs and DEGs(B)between adjacent stages or tissues at the same stage The code names of samples are consistent with those in Fig. 1.
| 样品 | 功能 Function | 基因编号 | 基因功能注释 |
|---|---|---|---|
| Sample | Gene ID | Gene annotation | |
| MY vs. PR | 碳水化合物代谢相关 Related to carbohydrate metabolism | jgi.p|Lenedo1|1049584 | 糖苷水解酶家族10蛋白 |
| Glycoside hydrolase family 10 protein | |||
| jgi.p|Lenedo1|1114896 | 内切葡聚糖酶V类似蛋白 | ||
| Endoglucanase V-like protein | |||
| jgi.p|Lenedo1|1038064 | 糖苷水解酶家族61蛋白 | ||
| Glycoside hydrolase family 61 protein | |||
| jgi.p|Lenedo1|1159040 | 糖苷水解酶家族95蛋白 | ||
| Glycoside hydrolase family 95 protein | |||
| jgi.p|Lenedo1|1158159 | 糖苷水解酶家族10蛋白 | ||
| Glycoside hydrolase family 10 protein | |||
| jgi.p|Lenedo1|464359 | 糖苷水解酶 Glycoside hydrolase | ||
| jgi.p|Lenedo1|1073151 | 糖苷水解酶家族18蛋白 | ||
| Glycoside hydrolase family 18 protein | |||
| 环境因子感知及信号转导相关 Related to environmental factor perception and signaling | jgi.p|Lenedo1|1035471 | Rheb小单体GTPase Rheb small monomeric gtpase | |
| jgi.p|Lenedo1|1083501 | COP9信号小体复合亚基4 | ||
| COP9 signalosome complex subunit 4 | |||
| jgi.p|Lenedo1|557311 | 假定水通道蛋白4 Putative aquaporin 4 | ||
| 细胞壁相关 Related to cell wall | jgi.p|Lenedo1|1053705 | 扩展蛋白 Expansin | |
| 蛋白合成与降解相关 Related to protein synthesis and degradation | jgi.p|Lenedo1|1100937 | 酸性蛋白酶 Acid protease | |
| jgi.p|Lenedo1|1060434 | 40S核糖体蛋白 S7 40S ribosomal protein S7 | ||
| jgi.p|Lenedo1|607412 | 核糖体蛋白L4 Ribosomal protein L4 | ||
| jgi.p|Lenedo1|1042368 | 三肽基肽酶A Tripeptidyl peptidase A | ||
| jgi.p|Lenedo1|1039990 | 26S蛋白酶体亚基 P45 26S proteasome subunit P45 | ||
| jgi.p|Lenedo1|1092580 | 泛素结合酶E2 Ubiquitin-conjugating enzyme E2 | ||
| jgi.p|Lenedo1|81445 | 肽酶s41家族蛋白 Peptidase s41 family protein | ||
| jgi.p|Lenedo1|1033416 | 核糖体蛋白L28e Ribosomal protein L28e | ||
| 其他功能 Other functions | jgi.p|Lenedo1|1059083 | 细胞色素P450 Cytochrome P450 | |
| jgi.p|Lenedo1|1066578 | 细胞色素P450 Cytochrome p450 | ||
| PR vs. D1ST | jgi.p|Lenedo1|717494 | MFS转运蛋白 MFS general substrate transporter |
Table1 Part genes both differentially expressed and spliced at adjacent stages or different tissues at the same stage
| 样品 | 功能 Function | 基因编号 | 基因功能注释 |
|---|---|---|---|
| Sample | Gene ID | Gene annotation | |
| MY vs. PR | 碳水化合物代谢相关 Related to carbohydrate metabolism | jgi.p|Lenedo1|1049584 | 糖苷水解酶家族10蛋白 |
| Glycoside hydrolase family 10 protein | |||
| jgi.p|Lenedo1|1114896 | 内切葡聚糖酶V类似蛋白 | ||
| Endoglucanase V-like protein | |||
| jgi.p|Lenedo1|1038064 | 糖苷水解酶家族61蛋白 | ||
| Glycoside hydrolase family 61 protein | |||
| jgi.p|Lenedo1|1159040 | 糖苷水解酶家族95蛋白 | ||
| Glycoside hydrolase family 95 protein | |||
| jgi.p|Lenedo1|1158159 | 糖苷水解酶家族10蛋白 | ||
| Glycoside hydrolase family 10 protein | |||
| jgi.p|Lenedo1|464359 | 糖苷水解酶 Glycoside hydrolase | ||
| jgi.p|Lenedo1|1073151 | 糖苷水解酶家族18蛋白 | ||
| Glycoside hydrolase family 18 protein | |||
| 环境因子感知及信号转导相关 Related to environmental factor perception and signaling | jgi.p|Lenedo1|1035471 | Rheb小单体GTPase Rheb small monomeric gtpase | |
| jgi.p|Lenedo1|1083501 | COP9信号小体复合亚基4 | ||
| COP9 signalosome complex subunit 4 | |||
| jgi.p|Lenedo1|557311 | 假定水通道蛋白4 Putative aquaporin 4 | ||
| 细胞壁相关 Related to cell wall | jgi.p|Lenedo1|1053705 | 扩展蛋白 Expansin | |
| 蛋白合成与降解相关 Related to protein synthesis and degradation | jgi.p|Lenedo1|1100937 | 酸性蛋白酶 Acid protease | |
| jgi.p|Lenedo1|1060434 | 40S核糖体蛋白 S7 40S ribosomal protein S7 | ||
| jgi.p|Lenedo1|607412 | 核糖体蛋白L4 Ribosomal protein L4 | ||
| jgi.p|Lenedo1|1042368 | 三肽基肽酶A Tripeptidyl peptidase A | ||
| jgi.p|Lenedo1|1039990 | 26S蛋白酶体亚基 P45 26S proteasome subunit P45 | ||
| jgi.p|Lenedo1|1092580 | 泛素结合酶E2 Ubiquitin-conjugating enzyme E2 | ||
| jgi.p|Lenedo1|81445 | 肽酶s41家族蛋白 Peptidase s41 family protein | ||
| jgi.p|Lenedo1|1033416 | 核糖体蛋白L28e Ribosomal protein L28e | ||
| 其他功能 Other functions | jgi.p|Lenedo1|1059083 | 细胞色素P450 Cytochrome P450 | |
| jgi.p|Lenedo1|1066578 | 细胞色素P450 Cytochrome p450 | ||
| PR vs. D1ST | jgi.p|Lenedo1|717494 | MFS转运蛋白 MFS general substrate transporter |
| [1] |
Almási E, Sahu N, Krizsán K, Bálint B, Kovács G M, Kiss B, Cseklye J, Drula E, Henrissat B, Nagy I, Chovatia M, Adam C, LaButti K, Lipzen A, Riley R, Grigoriev I V, Nagy L G. 2019. Comparative genomics reveals unique wood-decay strategies and fruiting body development in the Schizophyllaceae. New Phytologist, 224 (2):902-915.
doi: 10.1111/nph.16032 pmid: 31257601 |
| [2] |
Anders S, Pyl P T, Huber W. 2015. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics, 31 (2):166-169.
doi: 10.1093/bioinformatics/btu638 pmid: 25260700 |
| [3] |
Arima T, Yamamoto M, Hirata A, Kawano S, Kamada T. 2004. The eln3 gene involved in fruiting body morphogenesis of Coprinus cinereus encodes a putative membrane protein with a general glycosyltransferase domain. Fungal Genetics and Biology, 41 (8):805-812.
doi: 10.1016/j.fgb.2004.04.003 URL |
| [4] |
Attaran Dowom S, Rezaeian S, Pourianfar H R. 2019. Agronomic and environmental factors affecting cultivation of the winter mushroom or enokitake:achievements and prospects. Applied Microbiology and Biotechnology, 103 (6):2469-2481.
doi: 10.1007/s00253-019-09652-y URL |
| [5] |
Bahn Y S, Mühlschlegel F A. 2006. CO2 sensing in fungi and beyond. Current Opinion in Microbiology, 9 (6):572-578.
doi: 10.1016/j.mib.2006.09.003 URL |
| [6] |
Bolger A M, Lohse M, Usadel B. 2014. Trimmomatic:a flexible trimmer for Illumina sequence data. Bioinformatics, 30 (15):2114-2120.
doi: 10.1093/bioinformatics/btu170 URL |
| [7] |
Boulianne R P, Liu Y, Aebi M., Lu B, Kües U. 2000. Fruiting body development in Coprinus cinereus:regulated expression of two galectins secreted by a non-classical pathway. Microbiology, 146:1841-1853.
doi: 10.1099/00221287-146-8-1841 URL |
| [8] |
Cary J W, Harris-Coward P Y, Ehrlich K C, Di Mavungu J D, Malysheva S V, De Saeger S, Dowd P F, Shantappa S, Martens S L, Calvo A M. 2014. Functional characterization of a veA-dependent polyketide synthase gene in Aspergillus flavus necessary for the synthesis of asparasone,a sclerotium-specific pigment. Fungal Genetics and Biology, 64:25-35.
doi: 10.1016/j.fgb.2014.01.001 URL |
| [9] |
Cheawchanlertfa P, Cheevadhanarak S, Tanticharoen M, Maresca B, Laoteng K. 2011. Up-regulated expression of desaturase genes of Mucor rouxii in response to low temperature associates with pre-existing cellular fatty acid constituents. Molecular Biology Reports, 38 (5):3455-3462.
doi: 10.1007/s11033-010-0455-x pmid: 21104442 |
| [10] |
Chen C, Chen H, Zhang Y, Thomas H R, Frank M H, He Y, Xia R. 2020. TBtools:an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 13 (8):1194-1202.
doi: 10.1016/j.molp.2020.06.009 URL |
| [11] |
Eisenman H C, Casadevall A. 2012. Synthesis and assembly of fungal melanin. Applied Microbiology and Biotechnology, 93 (3):931-940.
doi: 10.1007/s00253-011-3777-2 pmid: 22173481 |
| [12] |
Ernst J, Bar-Joseph Z. 2006. STEM:a tool for the analysis of short time series gene expression data. BMC Bioinformatics, 7:191.
doi: 10.1186/1471-2105-7-191 URL |
| [13] |
Fang S M, Hou X, Qiu K H, He R, Feng X S, Liang X L. 2020. The occurrence and function of alternative splicing in fungi. Fungal Biology Reviews, 34 (4):178-188.
doi: 10.1016/j.fbr.2020.10.001 URL |
| [14] |
Gehrmann T, Pelkmans J F, Lugones L G, Wösten H A B, Abeel T, Reinders M J T. 2016. Schizophyllum commune has an extensive and functional alternative splicing repertoire. Scientific Reports, 6 (1):33640.
doi: 10.1038/srep33640 URL |
| [15] |
Grützmann K, Szafranski K, Pohl M, Voigt K, Petzold A, Schuster S. 2014. Fungal alternative splicing is associated with multicellular complexity and virulence:a genome-wide multi-species study. DNA Research, 21 (1):27-39.
doi: 10.1093/dnares/dst038 pmid: 24122896 |
| [16] |
Huang X, Zhang R, Qiu Y, Wu H, Xiang Q, Yu X, Zhao K, Zhang X, Chen Q, Penttinen P, Gu Y. 2020. RNA-seq profiling showed divergent carbohydrate-active enzymes(cazymes)expression patterns in Lentinula edodes at brown film formation stage under blue light induction. Frontiers in Microbiology, 11:1044.
doi: 10.3389/fmicb.2020.01044 URL |
| [17] | Hughes B T, Espenshade P J. 2008. Oxygen-regulated degradation of fission yeast SREBP by Ofd1,a prolyl hydroxylase family member. EMBO J, 27 (10):1491-1501. |
| [18] |
Kamada T, Sano H, Nakazawa T, Nakahori K. 2010. Regulation of fruiting body photomorphogenesis in Coprinopsis cinerea. Fungal Genetics and Biology, 47:917-921.
doi: 10.1016/j.fgb.2010.05.003 URL |
| [19] |
Kim D, Langmead B, Salzberg S L. 2015. HISAT:a fast spliced aligner with low memory requirements. Nature Methods, 12 (4):357-360.
doi: 10.1038/NMETH.3317 |
| [20] | Kim J Y, Kim D Y, Park Y J, Jang M J. 2020. Transcriptome analysis of the edible mushroom Lentinula edodes in response to blue light. PLoS ONE, 15 (3):e0230680. |
| [21] | Krizsán K, Almási E, Merényi Z, Sahu N, Virágh M, Kószó T, Mondo S, Kiss B, Bálint B, Kües U, Barry K, Cseklye J, Hegedüs B, Henrissat B, Johnson J, Lipzen A, Ohm R A, Nagy I, Pangilinan J, Yan J, Xiong Y, Grigoriev I V, Hibbett D S, Nagy L G. 2019. Transcriptomic atlas of mushroom development reveals conserved genes behind complex multicellularity in fungi. Proceedings of the National Academy of the Sciences of the United States of America, 116 (15):7409-7418. |
| [22] |
Langfelder P, Horvath S. 2008. WGCNA:an R package for weighted correlation network analysis. BMC Bioinformatics, 9:559.
doi: 10.1186/1471-2105-9-559 pmid: 19114008 |
| [23] |
Luan R, Liang Y, Chen Y, Liu H, Jiang S, Che T, Wong B, Sun H. 2010. Opposing developmental functions of Agrocybe aegerita galectin(AAL) during mycelia differentiation. Fungal Biology, 114 (8):599-608.
doi: 10.1016/j.funbio.2010.05.001 URL |
| [24] |
Manachère G. 1980. Conditions essential for controlled fruiting of macromycetes - a review. Transactions of the British Mycological Society, 75 (2):255-270.
doi: 10.1016/S0007-1536(80)80088-X URL |
| [25] |
Muraguchi H, Fujita T, Kishibe Y, Konno K, Ueda N, Nakahori K, Yanagi S O, Kamada T. 2008. The exp1 gene essential for pileus expansion and autolysis of the inky cap mushroom Coprinopsis cinerea(Coprinus cinereus)encodes an HMG protein. Fungal Genetics and Biology, 45 (6):890-896.
doi: 10.1016/j.fgb.2007.11.004 pmid: 18164224 |
| [26] |
Muraguchi H, Kamada T. 1998. The ich1 gene of the mushroom Coprinus cinereus is essential for pileus formation in fruiting. Development, 125 (16):3133-3141.
pmid: 9671586 |
| [27] |
Muraguchi H, Kamada T. 2000. A mutation in the eln2 gene encoding a cytochrome P 450 of Coprinus cinereus affects mushroom morphogenesis. Fungal Genetics and Biology, 29 (1):49-59.
pmid: 10779399 |
| [28] | Muraguchi H, Umezawa K, Niikura M, Yoshida M, Kozaki T, Ishii K, Sakai K, Shimizu M, Nakahori K, Sakamoto Y, Choi C, Ngan C Y, Lindquist E, Lipzen A, Tritt A, Haridas S, Barry K, Grigoriev I V, Pukkila P J. 2015. Strand-specific RNA-seq analyses of fruiting body development in Coprinopsis cinerea. PLoS ONE, 10 (10):e0141586. |
| [29] | Nagy L G, Kovács G M, Krizsán K. 2018. Complex multicellularity in fungi:evolutionary convergence,single origin,or both?Biological Reviews of the Cambridge Philosophical Society, 93 (4):1778-1794. |
| [30] |
Nehls U, Dietz S. 2014. Fungal aquaporins:cellular functions and ecophysiological perspectives. Applied Microbiology and Biotechnology, 98 (8):8835-8851.
doi: 10.1007/s00253-014-6049-0 URL |
| [31] |
Ohm R A, Aerts D, Wösten H A B, Lugones L G. 2013. The blue light receptor complex WC-1/ 2 of Schizophyllum commune is involved in mushroom formation and protection against phototoxicity. Environmental Microbiology, 15 (3):943-955.
doi: 10.1111/j.1462-2920.2012.02878.x URL |
| [32] |
Ohm R A, de Jong J F, Lugones L G, Aerts A, Kothe E, Stajich J E, de Vries R P, Record E, Levasseur A, Baker S E, Bartholomew K A, Coutinho P M, Erdmann S, Fowler T J, Gathman A C, Lombard V, Henrissat B, Knabe N, Kues U, Lilly W W, Lindquist E, Lucas S, Magnuson J K, Piumi F, Raudaskoski M, Salamov A, Schmutz J, Schwarze F W M R, vanKuyk P A, Horton J S, Grigoriev I V, Wösten H A B. 2010. Genome sequence of the model mushroom Schizophyllum commune. Nature Biotechnology, 28 (9):957-963.
doi: 10.1038/nbt.1643 |
| [33] | Pe P P W, Naing A H, Chung M Y, Park K I, Kim C K. 2019. The role of antifreeze proteins in the regulation of genes involved in the response of Hosta capitata to cold. 3 Biotech, 9 (9):335. |
| [34] | Pelkmans J F, Lugones L G, Wösten H A B. 2016. Fruiting body formation in basidiomycetes//Wendland J. Growth,differentiation and sexuality. The mycota(A comprehensive treatise on fungi as experimental systems for basic and applied research),Vol 1. Switzerland:Springer:387-405. |
| [35] |
Pelkmans J F, Patil M B, Gehrmann T, Reinders M J T, Wösten H A B, Lugones L G. 2017. Transcription factors of Schizophyllum commune involved in mushroom formation and modulation of vegetative growth. Scientific Reports, 7:310.
doi: 10.1038/s41598-017-00483-3 pmid: 28331193 |
| [36] |
Perez Di Giorgio J, Soto G, Alleva K, Jozefkowicz C, Amodeo G, Muschietti J P, Ayub N D. 2014. Prediction of aquaporin function by integrating evolutionary and functional analyses. The Journal of Membrane Biology, 247 (2):107-125.
doi: 10.1007/s00232-013-9618-8 URL |
| [37] | Pöggeler S, Nowrousian M, Teichert I, Beier A, Kück U. 2018. Fruiting-body development in ascomycetes//Kües U,Fischer R. Growth,differentiation and sexuality. The mycota(A comprehensive treatise on fungi as experimental systems for basic and applied research),Vol 1. Berlin:Springer:325-355. |
| [38] |
Purschwitz J, Muller S, Kastner C, Schoser M, Haas H, Espeso E A, Atoui A, Calvo A M, Fischer R. 2008. Functional and physical interaction of blue- and red-light sensors in Aspergillus nidulans. Current Biology, 18 (4):255-259
doi: 10.1016/j.cub.2008.01.061 pmid: 18291652 |
| [39] | Rio D C, Ares M, Hannon G J, Nilsen T W. 2010. Purification of RNA using TRIzol(TRI Reagent). Cold Spring Harb Protoc,doI: 10.1101/pdb.prot5439. |
| [40] |
Robinson C H. 2001. Cold adaptation in Arctic and Antarctic fungi. New Phytologist, 151 (2):341-353.
doi: 10.1046/j.1469-8137.2001.00177.x URL |
| [41] |
Robinson M D, McCarthy D J, Smyth G K. 2010. edgeR:a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26 (1):139-140.
doi: 10.1093/bioinformatics/btp616 pmid: 19910308 |
| [42] | Royse D J, Baars J, Tan Q. 2017. Current overview of mushroom production in the world:technology and applications//Diego C Z,Pardo-Giménez A. Edible and medicinal mushrooms:technology and applications. Hoboken:Wiley:5-13. |
| [43] |
Sakamoto Y, Minato K, Nagai M, Mizuno M, Sato T. 2005. Characterization of the Lentinula edodes exg2 gene encoding a lentinan-degrading exo-β-1,3-glucanase. Current Genetics, 48 (3):195-203.
pmid: 16133343 |
| [44] | Sakamoto Y, Nakade K, Sato S, Yoshida K, Miyazaki K, Natsume S, Konno N. 2017. Lentinula edodes genome survey and postharvest transcriptome analysis. Applied and Environmental Microbiology, 83 (10):e02990-16. |
| [45] |
Sano H, Kaneko S, Sakamoto Y, Sato T, Shishido K. 2009. The basidiomycetous mushroom Lentinula edodes white collar-2 homolog PHRB,a partner of putative blue-light photoreceptor PHRA,binds to a specific site in the promoter region of the L. edodes tyrosinase gene. Fungal Genetics and Biology, 46 (4):333-341.
doi: 10.1016/j.fgb.2009.01.001 URL |
| [46] |
Shimoda C, Uehira M, Kishida M, Fujioka H, Iino Y, Watanabe Y, Yamamoto M. 1987. Cloning and analysis of transcription of the mei2 gene responsible for initiation of meiosis in the fission yeast Schizosaccharomyces pombe. Journal of Bacteriology, 169 (1):93-96.
pmid: 3025190 |
| [47] |
Showalter A M. 1993. Structure and function of plant cell wall proteins. Plant Cell, 5 (1):9-23.
pmid: 8439747 |
| [48] | Song H Y, Kim D H, Kim J M. 2018. Comparative transcriptome analysis of dikaryotic mycelia and mature fruiting bodies in the edible mushroom Lentinula edodes. Scientific Reports, 8 (1):8983. |
| [49] | Song Linli, Xing Xiaoke, Guo Shunxing. 2018. Morphological process and regulation mechanisms of fruiting body differentiation in macrofungi:a review. Mycosystema, 37:671-684. (in Chinese) |
| 宋林丽, 邢晓科, 郭顺星. 2018. 大型真菌子实体发生的形态学过程及调控机制. 菌物学报, 37:671-684. | |
| [50] |
Tang L H, Jian H H, Song C Y, Bao D P, Shang X D, Wu D Q, Tan Q, Zhang X H. 2013. Transcriptome analysis of candidate genes and signaling pathways associated with light-induced brown film formation in Lentinula edodes. Applied Microbiology and Biotechnology, 97 (11):4977-4989.
doi: 10.1007/s00253-013-4832-y URL |
| [51] |
Tao Y X, Chen R L, Yan J J, Long Y, Tong Z J, Song H B, Xie B G. 2019. A hydrophobin gene,Hyd9,plays an important role in the formation of aerial hyphae and primordia in Flammulina filiformis. Gene, 706 (20):84-90.
doi: 10.1016/j.gene.2019.04.067 URL |
| [52] |
Tao Y X, Xie B G, Yang Z Y, Chen Z H, Chen B Z, Deng Y J, Jiang Y J, Peer A F. 2013. Identification and expression analysis of a new glycoside hydrolase family 55 exo-β-1,3-glucanase-encoding gene in Volvariella volvacea suggests a role in fruiting body development. Gene, 527 (1):154-160.
doi: 10.1016/j.gene.2013.05.071 URL |
| [53] |
Turner E M. 1977. Development of excised sporocarps of Agaricus bisporus and its control by CO2. Transactions of the British Mycological Society, 69 (2):183-186.
doi: 10.1016/S0007-1536(77)80035-1 URL |
| [54] |
Vetchinkina E, Kupryashina M, Gorshkov V, Ageeva M, Gogolev Y, Nikitina V. 2017. Alteration in the ultrastructural morphology of mycelial hyphae and the dynamics of transcriptional activity of lytic enzyme genes during basidiomycete morphogenesis. Journal of Microbiology, 55 (4):280-288.
doi: 10.1007/s12275-017-6320-z pmid: 28124773 |
| [55] |
Wang Q, Guo M, Xu R, Zhang J, Bian Y, Xiao Y. 2019. Transcriptional changes on blight fruiting body of Flammulina velutipes caused by two new bacterial pathogens. Frontiers in Microbiology, 10:2845.
doi: 10.3389/fmicb.2019.02845 URL |
| [56] |
Wang Y, Zeng X, Liu W. 2018. De novo transcriptomic analysis during Lentinula edodes fruiting body growth. Gene, 641:326-334.
doi: 10.1016/j.gene.2017.10.061 URL |
| [57] | Wang Zhuo ren, Liu Qi yan, Xiao Yang, Wu Qian, Bian Yin bing. 2010. Grey correlational and ISSR analyses of Lentinula edodes hybrids. Mycosystema, 29 (2):267-272. (in Chinese) |
| 王卓仁, 刘启燕, 肖扬, 吴茜, 边银丙. 2010. 香菇单孢杂交子代群体灰色关联度和ISSR分析. 菌物学报, 29 (2):267-272. | |
| [58] |
Wösten H A B, van Wetter M A, Lugones L G, van der Mei H C, Busscher H J, Wessels J G H. 1999. How a fungus escapes the water to grow into the air. Current Biology, 9 (2):85-88.
pmid: 10021365 |
| [59] |
Wu W, Zong J, Wei N, Cheng J, Zhou X, Cheng Y, Chen D, Guo Q, Zhang B, Feng Y. 2018. CASH:a constructing comprehensive splice site method for detecting alternative splicing events. Briefings in Bioinformatics, 19 (5):905-917.
doi: 10.1093/bib/bbx034 URL |
| [60] |
Yoo S I, Lee H Y, Markkandan K, Moon S, Ahn Y J, Ji S, Ko J, Kim S J, Ryu H, Hong C P. 2019. Comparative transcriptome analysis identified candidate genes involved in mycelium browning in Lentinula edodes. BMC Genomics, 20 (1):121.
doi: 10.1186/s12864-019-5509-4 URL |
| [61] |
Yu G, Wang L G, Han Y, He Q Y. 2012. clusterProfiler:an R package for comparing biological themes among gene clusters. Omics-A Journal of Integrative Biology, 16 (5):284-287.
doi: 10.1089/omi.2011.0118 URL |
| [62] |
Zerai D B, Fitzsimmons K M,and Collier R J. 2010. Transcriptional response of delta-9-desaturase gene to acute and chronic cold stress in Nile Tilapia,Oreochromis niloticus. Journal of the World Aquaculture Society, 41 (5):800-806.
doi: 10.1111/j.1749-7345.2010.00422.x URL |
| [63] | Zhang J C, Shen N, Li C, Xiang X J, Liu G L, Gui Y, Patev S, Hibbett D S, Barry K, Andreopoulos W, Lipzen A, Riley R, He G F, Yan M, Grigoriev I V, Kwan H S, Cheung M K, Bian Y B, Xiao Y. 2021. Population genomics provides insights into the genetic basis of adaptive evolution in the mushroom-forming fungus Lentinula edodes. Journal of Advanced Research,doi: 10.1016/j.jare.2021.09.008. |
| [64] | Zhao Chunqiao, Xie Fang, Wu Pingmin. 2021. Advance in researching the effects of environmental factors on sexual development of edible fungus. Chinese Agricultural Science Bulletin, 30:87-92. (in Chinese) |
| 赵春巧, 谢放, 吴萍民. 2014. 环境因素对食用菌有性发育影响的研究进展. 中国农学通报, 30:87-92. | |
| [65] | Zhou J, Kang L, Liu C, Niu X, Wang X, Liu H, Zhang W, Liu Z, Latge J P, Yuan S. 2019. Chitinases play a key role in stipe cell wall extension in the mushroom Coprinopsis cinerea. Applied and Environmental Microbiology, 85 (15):e00532-19. |
| [1] | JIANG Yu, TU Xunliang, and HE Junrong. Analysis of Differential Expression Genes in Leaves of Leaf Color Mutant of Chinese Orchid [J]. Acta Horticulturae Sinica, 2023, 50(2): 371-381. |
| [2] | ZHAO Xueyan, WANG Qi, WANG Li, WANG Fangyuan, WANG Qing, LI Yan. Comparative Transcriptome Analysis of Differential Expression in Different Tissues of Corydalis yanhusuo [J]. Acta Horticulturae Sinica, 2023, 50(1): 177-187. |
| [3] | ZHOU Xuzixin, YANG Wei, MAO Meiqin, XUE Yanbin, MA Jun. Identification of Pigment Components and Key Genes in Carotenoid Pathway in Mutants of Chimeric Ananas comosus var. bracteatus [J]. Acta Horticulturae Sinica, 2022, 49(5): 1081-1091. |
| [4] | XIA Ming, LI Jingwei, LUO Zhangrui, ZU Guidong, WANG Ya, ZHANG Wanping. Research on Mechanism of Exogenous Melatonin Effects on Radish Growth and Resistence to Alternaria brassicae [J]. Acta Horticulturae Sinica, 2022, 49(3): 548-560. |
| [5] | DENG Jiao, SU Mengyue, LIU Xuelian, OU Kefang, HU Zhengrong, YANG Pingfang. Transcriptome Analysis Revealed the Formation Mechanism of Floral Color of Lotus‘Dasajin’with Bicolor Petal [J]. Acta Horticulturae Sinica, 2022, 49(2): 365-377. |
| [6] | QIAO Jun, WANG Liying, LIU Jing, LI Suweng. Expression Analysis of Genes Related to Photosensitive Color Under the Caylx in Eggplant Based on Transcriptome Sequencing [J]. Acta Horticulturae Sinica, 2022, 49(11): 2347-2356. |
| [7] | WANG Ronghua, WANG Shubin, LIU Shuantao, LI Qiaoyun, ZHANG Zhigang, WANG Lihua, ZHAO Zhizhong. Transcriptome Analysis of Waxy Near-isogenic Lines in Chinese Cabbage Floral Axis [J]. Acta Horticulturae Sinica, 2022, 49(1): 62-72. |
| [8] | XU Hongxia, ZHOU Huifen, LI Xiaoying, JIANG Luhua, CHEN Junwei. Comparative Transcriptome Analysis of Different Developmental Stages of Flowers and Fruits in Loquat Under Low Temperature Stress [J]. Acta Horticulturae Sinica, 2021, 48(9): 1680-1694. |
| [9] | LIU Jianfeng, SUN Ying, WEI Heng, HE Hongli, ZHANG Xingzheng, CHENG Yunqing. Analysis and Identification of circRNAs of Hazel Ovule at Different Developmental Stages [J]. Acta Horticulturae Sinica, 2021, 48(6): 1053-1066. |
| [10] | LAN Liming, LUO Changguo, WANG Sanhong. Analysis of Resistance Mechanism to Powdery Mildew Based on Transcriptome Sequencing in Malus hupehensis [J]. Acta Horticulturae Sinica, 2021, 48(5): 860-872. |
| [11] | YU Shangqi, ZHANG Rui, GUO Zhongzhong, SONG Yan, FU Jiazhi, WU Pengyu, MA Zhihao. Dynamic Changes of Auxin and Analysis of Differentially Expressed Genes in Walnut Endocarp During Hardening [J]. Acta Horticulturae Sinica, 2021, 48(3): 487-504. |
| [12] | CHEN Chen, LIU Chenghui, SHI Lijia, JIANG Aili, HU Wenzhong. An Analysis of Alternative Splicing Events During Browning Inhibition of Fresh-cut Apples by Hydrogen Sulfide Treatment [J]. Acta Horticulturae Sinica, 2021, 48(11): 2121-2132. |
| [13] | GUO Jun1,2,ZHU Jie1,2,XIE Shangqian1,3,ZHANG Ye1,2,YE Beilei1,2,ZHENG Liyan2,and LING Peng1,3,* . Development of SSR Molecular Markers Based on Transcriptome and Analysis of Genetic Relationship of Germplasm Resources in Avocado [J]. ACTA HORTICULTURAE SINICA, 2020, 47(8): 1552-1564. |
| [14] | XU Junxu1,LI Qingzhu1,LI Ye2,YANG Liuyan1,LI Xin1,WANG Zhen1,and ZHANG Yongchun1,*. Differential Expression of Genes Related to Endogenous Hormone During Bulb Development in Lycoris radiata [J]. ACTA HORTICULTURAE SINICA, 2020, 47(6): 1126-1140. |
| [15] | JU Lixiang,LEI Xin,ZHAO Chengzhi,SHU Huangying,WANG Zhiwei,and CHENG Shanhan*. Identification of MYB Family Genes and Its Relationship with Pungency of Pepper [J]. ACTA HORTICULTURAE SINICA, 2020, 47(5): 875-892. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd