
Acta Horticulturae Sinica ›› 2022, Vol. 49 ›› Issue (4): 739-748.doi: 10.16420/j.issn.0513-353x.2021-0179
• Research Papers • Previous Articles Next Articles
GAO Weilin1, ZHANG Liman1, XUE Chaoling1, ZHANG Yao1, LIU Mengjun2, ZHAO Jin1,*( )
)
Received:2021-11-18
Revised:2022-01-18
Online:2022-04-25
Published:2022-04-24
Contact:
ZHAO Jin
E-mail:zhaojinbd@126.com
CLC Number:
GAO Weilin, ZHANG Liman, XUE Chaoling, ZHANG Yao, LIU Mengjun, ZHAO Jin. Expression of E-type MADS-box Genes in Flower and Fruits and Protein Interaction Analysis in Chinese Jujube[J]. Acta Horticulturae Sinica, 2022, 49(4): 739-748.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2021-0179
| 用途 Usage | 基因名称 Gene name | 正向序列(5′-3′) Forward primer sequence | 反向序列(5′-3′) Reverse primer sequence |
|---|---|---|---|
| q-RT-PCR Quantitative real-time PCR | ZjSEP1/2 | GGGGAGAGGTAAGGTGGAG | GGTTGGAGAAGATGATAAGAGC |
| ZjSEP3 | GGGGAAGACCTTGGACCTTT | AGCAAGTGTTCCTTGCGTTG | |
| ZjSEP4 | GGGGTTCTTCCAGGCCTTAG | ACTTGCTGGGCATGACTTGT | |
| 内参 Primers for internal control | ZjACT | AGCCTTCCTGCCAACGAGT | TTGCTTCTCACCCTTGATGC |
| 酵母双杂交 Yeast two- Hybrid(Y2H) | ZjBDZjSEP1/2 | ATGGCCATGGAGGCCGATGGGGAGAGGTAAGGTGGA | GGATCCCCGGGAATTTCAAAGCATCCACCCAGGG |
| ZjBDZjSEP3 | GGATCCCCGGGAATTTCAAAGCATCCACCCAGGG | GGATCCCCGGGAATTTCATGGTAACCATCCTGGCA | |
| ZjBDZjSEP4 | ATGGCCATGGAGGCCGATGGGGAGGGGTAGAGTTGA | GGATCCCCGGGAATTTCAAAGCATCCATCCAGGAA | |
| ZjADZjSEP1/2 | CCATGGAGGCCAGTGATGGGGAGAGGTAAGGTGGA | CACCCGGGTGGAATTTCAAAGCATCCACCCAGGG | |
| ZjADZjSEP3 | CCATGGAGGCCAGTGATGGGAAGAGGTAGAGTGGA | CACCCGGGTGGAATTTCATGGTAACCATCCTGGCA | |
| ZjADZjSEP4 | CCATGGAGGCCAGTGATGGGGAGGGGTAGAGTTGA | CACCCGGGTGGAATTTCAAAGCATCCATCCAGGAA | |
| ZjADZjMADS46 | CCATGGAGGCCAGTGATGGAGTTCCCAAATCAAGCA | CACCCGGGTGGAATTTCAAACAAGTTGGAGAGCTG | |
| ZjADMADS24 | CCATGGAGGCCAGTGATGGGGAGGGGAAGGGTTC | CACCCGGGTGGAATTGCATCCAAGCGGGCAGGAG | |
| 亚细胞定位 Subcellular localization | ZjSEP1/2 | AGCTCGGGTACCCGGGATGGGGAGAGGTAAGGTGGA | TCTAGAGGATCAAGCATCCACCCAGGG |
| ZjSEP3 | AGCTCGGGTACCCGGGATGCTCAAAACACTTGAGA | TCTAGAGGATCTGGTAACCATCCTGGCAT | |
| ZjSEP4 | AGCTCGGGTACCCGGGATGGGGAGGGGTAGAGTTGA | TCTAGAGGATCAAGCATCCATCCAGGAA |
Table 1 Primer names and sequences used in this study
| 用途 Usage | 基因名称 Gene name | 正向序列(5′-3′) Forward primer sequence | 反向序列(5′-3′) Reverse primer sequence |
|---|---|---|---|
| q-RT-PCR Quantitative real-time PCR | ZjSEP1/2 | GGGGAGAGGTAAGGTGGAG | GGTTGGAGAAGATGATAAGAGC |
| ZjSEP3 | GGGGAAGACCTTGGACCTTT | AGCAAGTGTTCCTTGCGTTG | |
| ZjSEP4 | GGGGTTCTTCCAGGCCTTAG | ACTTGCTGGGCATGACTTGT | |
| 内参 Primers for internal control | ZjACT | AGCCTTCCTGCCAACGAGT | TTGCTTCTCACCCTTGATGC |
| 酵母双杂交 Yeast two- Hybrid(Y2H) | ZjBDZjSEP1/2 | ATGGCCATGGAGGCCGATGGGGAGAGGTAAGGTGGA | GGATCCCCGGGAATTTCAAAGCATCCACCCAGGG |
| ZjBDZjSEP3 | GGATCCCCGGGAATTTCAAAGCATCCACCCAGGG | GGATCCCCGGGAATTTCATGGTAACCATCCTGGCA | |
| ZjBDZjSEP4 | ATGGCCATGGAGGCCGATGGGGAGGGGTAGAGTTGA | GGATCCCCGGGAATTTCAAAGCATCCATCCAGGAA | |
| ZjADZjSEP1/2 | CCATGGAGGCCAGTGATGGGGAGAGGTAAGGTGGA | CACCCGGGTGGAATTTCAAAGCATCCACCCAGGG | |
| ZjADZjSEP3 | CCATGGAGGCCAGTGATGGGAAGAGGTAGAGTGGA | CACCCGGGTGGAATTTCATGGTAACCATCCTGGCA | |
| ZjADZjSEP4 | CCATGGAGGCCAGTGATGGGGAGGGGTAGAGTTGA | CACCCGGGTGGAATTTCAAAGCATCCATCCAGGAA | |
| ZjADZjMADS46 | CCATGGAGGCCAGTGATGGAGTTCCCAAATCAAGCA | CACCCGGGTGGAATTTCAAACAAGTTGGAGAGCTG | |
| ZjADMADS24 | CCATGGAGGCCAGTGATGGGGAGGGGAAGGGTTC | CACCCGGGTGGAATTGCATCCAAGCGGGCAGGAG | |
| 亚细胞定位 Subcellular localization | ZjSEP1/2 | AGCTCGGGTACCCGGGATGGGGAGAGGTAAGGTGGA | TCTAGAGGATCAAGCATCCACCCAGGG |
| ZjSEP3 | AGCTCGGGTACCCGGGATGCTCAAAACACTTGAGA | TCTAGAGGATCTGGTAACCATCCTGGCAT | |
| ZjSEP4 | AGCTCGGGTACCCGGGATGGGGAGGGGTAGAGTTGA | TCTAGAGGATCAAGCATCCATCCAGGAA |
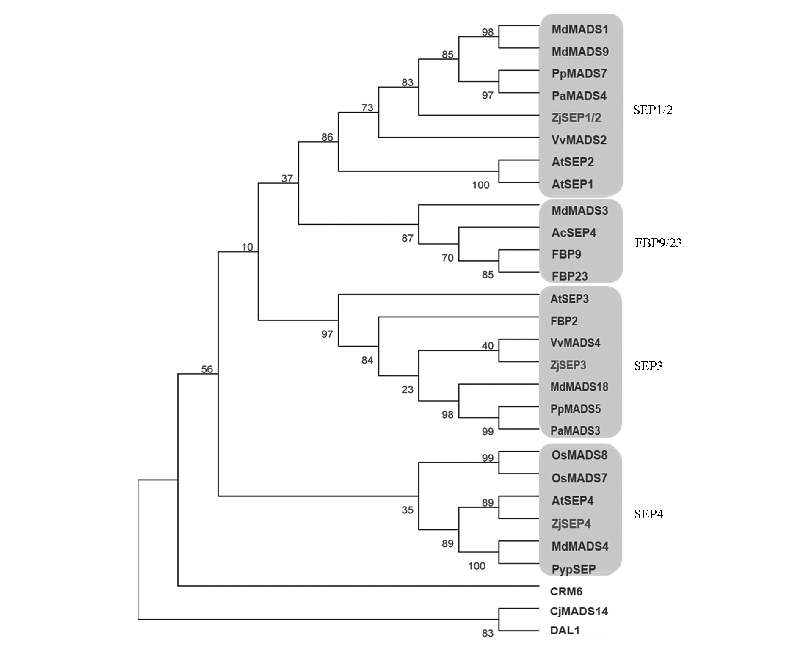
Fig. 1 Phylogenetic tree of E-type proteins from Chinese jujube and other species At:Arabidopsis thaliana;Md:Malus × domestica;Pp:Prunus persica;Pa:Prunus avium;Ac:Actinidia chinensis;Vv:Vitis vinifera;Pyp:Pyrus pyrifolia;Os:Oryza sativa;Cj:Cryptomeria japonica;DAL1:Picea abies;CRM6:Ceratopteris pteridoides;FBP:Petunia hybrida.

Fig. 2 Expression pattern of jujube E-type genes in flowers(A),fruit(B)and four-whorl floral organs(C) According to the Duncan’s multiple range test,the different letters above the column indicate the significant differences in flower,fruit at different development stages and four-wheel flower organs for each gene(P < 0.05).
| [1] |
Becker A, Theissen G. 2003. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Molecular Phylogenetics and Evolution, 29 (3):464-489.
doi: 10.1016/S1055-7903(03)00207-0 URL |
| [2] |
Coen E S, Meyerowitz E M. 1991. The war of the whorls:genetic interactions controlling flower development. Nature, 353 (6339):31-37.
doi: 10.1038/353031a0 URL |
| [3] |
Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky M F. 2004. The SEP 4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Current Biology, 14 (21):1935-1940.
pmid: 15530395 |
| [4] |
Günter Theißen. 2001. Development of floral organ identity:stories from the MADS house. Current Opinion in Plant Biology, 4 (1):75-85.
doi: 10.1016/S1369-5266(00)00139-4 URL |
| [5] |
Honma T, Goto K. 2001. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature, 409 (6819):525-529.
doi: 10.1038/35054083 URL |
| [6] | Huang Fang. 2007. Cloning and functional analysis of flower development related genes in soybean(Glycine max L.)[Ph. D. Dissertation]. Nanjing: Nanjing Agricultural University. (in Chinese) |
| 黄方. 2007. 大豆花发育相关基因的克隆与功能研究[博士论文]. 南京: 南京农业大学. | |
| [7] |
Huang J, Zhang C, Zhao X, Fei Z, Wan K, Zhang Z, Pang X, Yin X, Bai Y, Sun X, Gao L, Li R, Zhang J, Li X. 2016. The jujube genome provides insights into genome evolution and the domestication of sweetness/acidity taste in fruit trees. PLoS Genetics, 12 (12):e1006433.
doi: 10.1371/journal.pgen.1006433 URL |
| [8] |
Immink R G, Tonaco I A, de Folter S, Shchennikova A, van Dijk A D, Busscher-Lange J, Borst J W, Angenent G C. 2009. SEPALLATA3:the‘glue’for MADS box transcription factor complex formation. Genome Biology, 10 (2):R24.
doi: 10.1186/gb-2009-10-2-r24 URL |
| [9] |
Ireland H S, Yao J L, Tomes S, Sutherland P W, Nieuwenhuizen N, Gunaseelan K, Winz R A, David K M, Schaffer R J. 2013. Apple SEPALLATA1/2-like genes control fruit flesh development and ripening. Plant Journal, 73 (6):1044-1056.
doi: 10.1111/tpj.12094 URL |
| [10] |
Kobayashi K, Yasuno N, Sato Y, Yoda M, Yamazaki R, Kimizu M, Yoshida H, Nagamura Y, Kyozuka J. 2012. Inflorescence meristem identity in rice is specified by overlapping functions of three AP 1 /FUL-like MADS box genes and PAP2,a SEPALLATA MADS box gene. Plant Cell, 24 (5):1848-1859.
doi: 10.1105/tpc.112.097105 URL |
| [11] |
Krizek B A, Meyerowitz E M. 1996. The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development, 122 (1):11-22.
pmid: 8565821 |
| [12] | Li Chengru, Dong Na, Li Xiaoping, Wu Shasha, Liu Zhongjian, Zhai Junwen. 2020. A review of MADS-box genes,the molecular regulatory genes for floral organ development in Orchidaceae. Acta Horticulturae Sinica, 47 (10):2047-2062. (in Chinese) |
| 李成儒, 董钠, 李笑平, 吴沙沙, 刘仲健, 翟俊文. 2020. 兰科植物花发育调控MADS-box基因家族研究进展. 园艺学报, 47 (10):2047-2062. | |
| [13] |
Liao Y T, Lin S S, Lin S J, Shen B N, Cheng H P, Lin C P. 2019. Structural insights into the interaction between phytoplasmal effector causing phyllody 1(PHYL1)and MADS transcription factor. Plant Journal, 100 (4):706-719.
doi: 10.1111/tpj.14463 URL |
| [14] |
Lin X, Wu F, Du X, Shi X, Liu Y, Liu S, Hu Y, Theissen G, Meng Z. 2014. The pleiotropic SEPALLATA-like gene OsMADS34 reveals that the‘empty glumes’of rice(Oryza sativa)spikelets are in fact rudimentary lemmas. New Phytologist, 202 (2):689-702.
doi: 10.1111/nph.12657 URL |
| [15] | Liu M J, Zhao J, Cai Q L, Liu G C, Wang J R, Zhao Z H, Liu P, Dai L, Yan G, Wang W J, Li X S, Chen Y, Sun Y D, Liu Z G, Lin M J, Xiao J, Chen Y Y, Li X F, Wu B, Ma Y, Jian J B, Yang W, Yuan Z, Sun X C, Wei Y L, Yu L L, Zhang C, Liao S G, He R J, Guang X M, Wang Z, Zhang Y Y, Luo L H. 2014. The complex jujube genome provides insights into fruit tree biology. Nature Communications, 28 (5):5315. |
| [16] | Liu Shinan, Qi Tiantian, Ma Jingjing, Ma Lyuyi, Lin Xinchun. 2016. Cloning and functional analysis of SEP-like gene from phyllostachys violascens. Journal of Nuclear Agricultural Sciences, 30 (8):1453-1459. (in Chinese) |
| 刘世男, 戚田田, 马晶晶, 马履一, 林新春. 2016. 雷竹SEP-like基因的克隆及功能分析. 核农学报, 30 (8):1453-1459. | |
| [17] |
Ma H, Yanofsky M F, Meyerowitz E M. 1991. AGL1AGL6,an Arabidopsis gene family with similarity to floral hometic and transcription factor genes. Genes and Development, 5 (3):484-495.
pmid: 1672119 |
| [18] | MacLean A M, Sugio A, Makarova O V, Findlay K C, Grieve V M. 2011. Phytoplasma effector SAP54 induces indeterminate leaf-like flower development in Arabidopsis plants. Plant Physiology 157:831-841. |
| [19] |
Maejima K, Iwai R, Himeno M, Komatsu K, Kitazawa Y, Fujita N, Ishikawa K, Fukuoka M, Minato N, Yamaji Y, Oshima K, Namba S. 2014. Recognition of floral homeotic MADS domain transcription factors by a phytoplasmal effector,phyllogen,induces phyllody. Plant Journal, 78 (4):541-554.
doi: 10.1111/tpj.12495 URL |
| [20] |
Maejima K, Kitazawa Y, Tomomitsu T, Yusa A, Neriya Y, Himeno M, Yamaji Y, Oshima K, Namba S. 2015. Degradation of class E MADs-domain transcription factors in Arabidopsis by a phytoplasmal effector,phyllogen. Plant Signaling and Behavior, 10 (8):e1042635.
doi: 10.1080/15592324.2015.1042635 URL |
| [21] |
Melzer R, Verelst W, Theissen G. 2009. The class E floral homeotic protein SEPALLATA 3 is sufficient to loop DNA in‘floral quartet’-like complexes in vitro. Nucleic Acids Research, 37 (1):144-157.
doi: 10.1093/nar/gkn900 URL |
| [22] |
Meng X W, Li Y, Yuan Y, Zhang Y, Li H T, Zhao J, Liu M J. 2020. The regulatory pathways of distinct flowering characteristics in Chinese jujube. Horticulture Research, 7 (1):13-19.
doi: 10.1038/s41438-019-0236-1 URL |
| [23] |
Ng M, Yanofsky M F. 2001. Function and evolution of the plant MADS box gene family. Nature Reviews Genetics, 2:186-195.
pmid: 11256070 |
| [24] |
Pelaz S, Ditta G S, Baumann E, Wisman E, Yanofsky M F. 2000. B and C floral organ identity functions require SEPALLATA MADS box genes. Nature, 405 (6783):200-203.
doi: 10.1038/35012103 URL |
| [25] |
Pelaz S, Tapia-López R, Alvarez-Buylla E R, Yanofsky M F. 2001. Conversion of leaves into petals in Arabidopsis. Current Biology, 11 (3):182-184.
pmid: 11231153 |
| [26] |
Seymour G B., Ryder C D, Cevik V, Hammond J P, Popovich A., King G J, Vrebalov J, Giovannoni J J, Manning K. 2011. A SEPALLATA gene is involved in the development and ripening of strawberry(Fragaria × ananassa Duch.)fruit,a non-climacteric tissue. Journal of Experimental Botany, 62 (3):1179-1188.
doi: 10.1093/jxb/erq360 pmid: 21115665 |
| [27] | Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6:molecular evolutionary genetics analysis Version 6.0. Molecular Biology & Evolution, 30 (12):2725-2729. |
| [28] | Teo Z W N, Zhou W, Shen L. 2019. Dissecting the function of MADS-box transcription factors in orchid reproductive development. Frontiers in Plant Science, 15 (10):1474. |
| [29] |
Xu K, Huang X, Wu M, Wang Y, Chang Y, Liu K, Zhang J, Zhang Y, Zhang F, Yi L, Li T, Wang R, Tan G, Li C. 2014a. A rapid,highly efficient and economical method of Agrobacterium-mediated in planta transient transformation in living onion epidermis. PLoS ONE, 9 (1):e83556.
doi: 10.1371/journal.pone.0083556 URL |
| [30] | Xu Y, Zhang L, Xie H, Zhang Y Q, Oliveira M M, Ma R C. 2008. Expression analysis and genetic mapping of three SEPALLATA -like genes from peach(Prunus persica(L.)Batsch). Tree Genetics & Genomes, 4 (4):693-703. |
| [31] |
Xu Z, Zhang Q, Sun L, Du D, Cheng T, Pan H, Yang W, Wang J. 2014b. Genome-wide identification,characterisation and expression analysis of the MADS-box gene family in Prunus mume. Molecular Genetics and Genomics, 289 (5):903-920.
doi: 10.1007/s00438-014-0863-z URL |
| [32] |
Zhang L M, Zhao J, Feng C F, Liu M J, Wang J R, Hu Y F. 2017. Genome-wide identification,characterization of the MADS-box gene family in Chinese jujube and their involvement in flower development. Scientific Reports, 7 (1):1025.
doi: 10.1038/s41598-017-01159-8 URL |
| [33] |
Zhou Y, Xu Z, Yong X, Ahmad S, Yang W, Cheng T, Wang J, Zhang Q. 2017. SEP-class genes in Prunus mume and their likely role in floral organ development. BMC Plant Biology, 17 (1):10.
doi: 10.1186/s12870-016-0954-6 URL |
| [34] | Zong Chengwen, Fang Jinggui, Tao Jianmin, Zhang Zhen, Qian Yaming. 2007. Cloning and sequence analysis of MADS-box family protein gene fragments from grape. Journal of Fruit Science, 25 (1):27-32. (in Chinese) |
| 宗成文, 房经贵, 陶建敏, 章镇, 钱亚明. 2007. 葡萄MADS-box家族基因保守片段的克隆与序列分析. 果树学报, 25 (1):27-32. |
| [1] | ZHAO Xueyan, WANG Qi, WANG Li, WANG Fangyuan, WANG Qing, LI Yan. Comparative Transcriptome Analysis of Differential Expression in Different Tissues of Corydalis yanhusuo [J]. Acta Horticulturae Sinica, 2023, 50(1): 177-187. |
| [2] | GAO Yanlong, WU Yuxia, ZHANG Zhongxing, WANG Shuangcheng, ZHANG Rui, ZHANG De, WANG Yanxiu. Bioinformatics Analysis of Apple ELO Gene Family and Its Expression Analysis Under Low Temperature Stress [J]. Acta Horticulturae Sinica, 2022, 49(8): 1621-1636. |
| [3] | QIU Ziwen, LIU Linmin, LIN Yongsheng, LIN Xiaojie, LI Yongyu, WU Shaohua, YANG Chao. Cloning and Functional Analysis of the MbEGS Gene from Melaleuca bracteata [J]. Acta Horticulturae Sinica, 2022, 49(8): 1747-1760. |
| [4] | ZHENG Lin, WANG Shuai, LIU Yunuo, DU Meixia, PENG Aihong, HE Yongrui, CHEN Shanchun, ZOU Xiuping. Gene Cloning and Expression Analysis of NAC Gene in Citrus in Response to Huanglongbing [J]. Acta Horticulturae Sinica, 2022, 49(7): 1441-1457. |
| [5] | MA Weifeng, LI Yanmei, MA Zonghuan, CHEN Baihong, MAO Juan. Identification of Apple POD Gene Family and Functional Analysis of MdPOD15 Gene [J]. Acta Horticulturae Sinica, 2022, 49(6): 1181-1199. |
| [6] | ZHANG Kai, MA Mingying, WANG Ping, LI Yi, JIN Yan, SHENG Ling, DENG Ziniu, MA Xianfeng. Identification of HSP20 Family Genes in Citrus and Their Expression in Pathogen Infection Responses Citrus Canker [J]. Acta Horticulturae Sinica, 2022, 49(6): 1213-1232. |
| [7] | LIANG Chen, SUN Ruyi, XIANG Rui, SUN Yimeng, SHI Xiaoxin, DU Guoqiang, WANG Li. Genome-wide Identification of Grape GRF Family and Expression Analysis [J]. Acta Horticulturae Sinica, 2022, 49(5): 995-1007. |
| [8] | XIAO Xuechen, LIU Mengyu, JIANG Mengqi, CHEN Yan, XUE Xiaodong, ZHOU Chengzhe, WU Xingjian, WU Junnan, GUO Yinsheng, YEH Kaiwen, LAI Zhongxiong, LIN Yuling. Whole-genome Identification and Expression Analysis of SNAT,ASMT and COMT Families of Melatonin Synthesis Pathway in Dimocarpus longan [J]. Acta Horticulturae Sinica, 2022, 49(5): 1031-1046. |
| [9] | LIU Mengyu, JIANG Mengqi, CHEN Yan, ZHANG Shuting, XUE Xiaodong, XIAO Xuechen, LAI Zhongxiong, LIN Yuling. Genome-wide Identification and Expression Analysis of GDSL Esterase/Lipase Genes in Dimocarpus longan [J]. Acta Horticulturae Sinica, 2022, 49(3): 597-612. |
| [10] | JIANG Cuicui, FANG Zhizhen, ZHOU Danrong, LIN Yanjuan, YE Xinfu. Identification and Expression Analysis of Sugar Transporter Family Genes in‘Furongli’(Prunus salicina) [J]. Acta Horticulturae Sinica, 2022, 49(2): 252-264. |
| [11] | WANG Zhiyu, CHANG Beibei, LIU Qi, CHENG Xiaofan, DU Xiaoyun, YU Xiaoli, SONG Laiqing, ZHAO Lingling. Study on Expression and Anthocyanin Accumulation of Solute Carrier Gene MdSLC35F2-like in Apple [J]. Acta Horticulturae Sinica, 2022, 49(11): 2293-2303. |
| [12] | HUANG Renwei, REN Yinghong, QI Weiliang, ZENG Rui, LIU Xinyu, DENG Binyan. Cloning of Mulberry MaERF105-Like Gene and Its Expression Under Drought Stress [J]. Acta Horticulturae Sinica, 2022, 49(11): 2439-2448. |
| [13] | LI Ying, MENG Xianwei, MA Zhihang, LIU Mengjun, ZHAO Jin. Identification and Expression Analysis of MicroRNA Families Associated with Phase Transition in Chinese Jujube [J]. Acta Horticulturae Sinica, 2022, 49(1): 23-40. |
| [14] | GUO Xuemin, WANG Xinrui, WANG Yingying, LI Zheng, MIAO Ningning, WANG Zhaojun. Anatomical Observation on the Tortuousness of Ziziphus jujuba var. tortuosa Branches [J]. Acta Horticulturae Sinica, 2021, 48(9): 1653-1664. |
| [15] | BIAN Shicun, LU Yani, XU Wujun, CHEN Boqing, WANG Guanglong, XIONG Aisheng. Garlic Circadian Clock Genes AsRVE1 and AsRVE2 and Their Expression Analysis Under Osmotic Stress [J]. Acta Horticulturae Sinica, 2021, 48(9): 1706-1716. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd