
Acta Horticulturae Sinica ›› 2022, Vol. 49 ›› Issue (2): 281-292.doi: 10.16420/j.issn.0513-353x.2021-0045
• Research Papers • Previous Articles Next Articles
SONG Fang, LI Zixuan, WANG Ce, WANG Zhijing, HE Ligang, JIANG Yingchun, WU Liming( ), BAI Fuxi(
), BAI Fuxi( )
)
Received:2021-11-09
Revised:2022-01-07
Online:2022-02-25
Published:2022-02-28
Contact:
WU Liming,BAI Fuxi
E-mail:wuliming2005@126.com;baifx@webmail.hzau.edu.cn
CLC Number:
SONG Fang, LI Zixuan, WANG Ce, WANG Zhijing, HE Ligang, JIANG Yingchun, WU Liming, BAI Fuxi. Cloning and Function Analysis of Mycorrhizal Signaling Receptor Protein Lysin Motif Receptor-like Kinases 2 Gene(LYK2)in Citrus[J]. Acta Horticulturae Sinica, 2022, 49(2): 281-292.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2021-0045
| 登录号 Accession number | 基因 Gene | 正向引物序列(5′-3′) Forward primer sequence | 反向引物序列(5′-3′) Reverse primer sequence |
|---|---|---|---|
| Cs1g15820.1 | PtrLYK1 | GCAGATTGCACTTGATGCTG | CTTTGCTCGAAAAGCACTGTC |
| Cs1g23560.1 | PtrLYK2 | CCACAATTGATGGGAAGGTC | ACTTCACCAGATTGGCATGG |
| Cs1g23580.1 | PtrLYK3 | TGTTGGACTTGCCACAACAG | ACGGTGTTGAAAGGAGGTTG |
| Cs2g02680.1 | PtrLYK4 | ATCCGATTGAGCTTGGTGAG | GGATGAAAACCAAGCCACTG |
| Cs2g20910.1 | PtrLYK5 | AGTCGATTGCGAGGAATGTG | CCCGGAATAAAGACCAAACC |
| Cs2g25010.1 | PtrLYK6 | GAACCCCAGTTATGTTCACAGG | AATTGTGGGCTCTCGTTGTC |
| Cs6g02700.1 | PtrLYK7 | TTTGAGTGTGGTGGTTGTGG | TGTTGTTGCAGCAAGTAGCC |
| Cs6g02710.1 | PtrLYK8 | CCATGGATCTTGAAGGGTTG | TCCTCTGGCACGGAAAATAC |
| Cs7g09310.1 | PtrLYK9 | TCACACGAAAGCTCGATACG | TTCCTCAAGCTTCACCAACC |
| Cs8g16260.1 | PtrLYK10 | CCCGATGATTTCTCAAGCAG | GGAGATCCCGAATTTCATCC |
| Cs9g08050.1 | PtrLYK11 | AATCCCAGTGCCTTTCCTTC | TTTGGCATGGATAGGTGGTC |
| orange1.1t04036.1 | PtrLYK12 | TGAAGTGGTGTTGGTGTTGG | CGATGGATACAATCCGTTGC |
| Cs1g05000.1 | β-actin | AGAACTATGAACTGCCTGATGGC | GCTTGGAGCAAGTGCTGTGATT |
Table 1 The primers used for qRT-PCR
| 登录号 Accession number | 基因 Gene | 正向引物序列(5′-3′) Forward primer sequence | 反向引物序列(5′-3′) Reverse primer sequence |
|---|---|---|---|
| Cs1g15820.1 | PtrLYK1 | GCAGATTGCACTTGATGCTG | CTTTGCTCGAAAAGCACTGTC |
| Cs1g23560.1 | PtrLYK2 | CCACAATTGATGGGAAGGTC | ACTTCACCAGATTGGCATGG |
| Cs1g23580.1 | PtrLYK3 | TGTTGGACTTGCCACAACAG | ACGGTGTTGAAAGGAGGTTG |
| Cs2g02680.1 | PtrLYK4 | ATCCGATTGAGCTTGGTGAG | GGATGAAAACCAAGCCACTG |
| Cs2g20910.1 | PtrLYK5 | AGTCGATTGCGAGGAATGTG | CCCGGAATAAAGACCAAACC |
| Cs2g25010.1 | PtrLYK6 | GAACCCCAGTTATGTTCACAGG | AATTGTGGGCTCTCGTTGTC |
| Cs6g02700.1 | PtrLYK7 | TTTGAGTGTGGTGGTTGTGG | TGTTGTTGCAGCAAGTAGCC |
| Cs6g02710.1 | PtrLYK8 | CCATGGATCTTGAAGGGTTG | TCCTCTGGCACGGAAAATAC |
| Cs7g09310.1 | PtrLYK9 | TCACACGAAAGCTCGATACG | TTCCTCAAGCTTCACCAACC |
| Cs8g16260.1 | PtrLYK10 | CCCGATGATTTCTCAAGCAG | GGAGATCCCGAATTTCATCC |
| Cs9g08050.1 | PtrLYK11 | AATCCCAGTGCCTTTCCTTC | TTTGGCATGGATAGGTGGTC |
| orange1.1t04036.1 | PtrLYK12 | TGAAGTGGTGTTGGTGTTGG | CGATGGATACAATCCGTTGC |
| Cs1g05000.1 | β-actin | AGAACTATGAACTGCCTGATGGC | GCTTGGAGCAAGTGCTGTGATT |
| 用途 Purpose | 引物名称 Primer name | 引物序列(5′-3′) Primer sequence |
|---|---|---|
| PtrLYK2 CDS序列扩增 Amplification of coding sequence of PtrLYK2 | PtrLYK2-F | ATGGCAATTTCTTCTCTTTCCT |
| PtrLYK2-R | GCGAGCTGTGACTGGACTAA | |
| PtrLYK2启动子序列扩增 Amplification of promoter sequence of PtrLYK2 | proPtrLYK2-F | GATAGAACACTTTGCGGTC |
| proPtrLYK2-R | TGGTACAAATGTTCTTTTGT |
Table 2 The primers used for cloning CDS and promoter sequence of PtrLYK2
| 用途 Purpose | 引物名称 Primer name | 引物序列(5′-3′) Primer sequence |
|---|---|---|
| PtrLYK2 CDS序列扩增 Amplification of coding sequence of PtrLYK2 | PtrLYK2-F | ATGGCAATTTCTTCTCTTTCCT |
| PtrLYK2-R | GCGAGCTGTGACTGGACTAA | |
| PtrLYK2启动子序列扩增 Amplification of promoter sequence of PtrLYK2 | proPtrLYK2-F | GATAGAACACTTTGCGGTC |
| proPtrLYK2-R | TGGTACAAATGTTCTTTTGT |

Fig. 3 The phylogenetic tree of LYK family genes Cs:Citrus sinensis;Sl:Solanum lycopersicum;At:Arabidopsis thaliana;Pa: Parasponia andersonii;Gm:Glycine max;Lj:Lotus japonicus;Os:Oryza sativa;Pt:Populus trichocarpa;Vv:Vitis vinifera;Mt:Medicago truncatula.
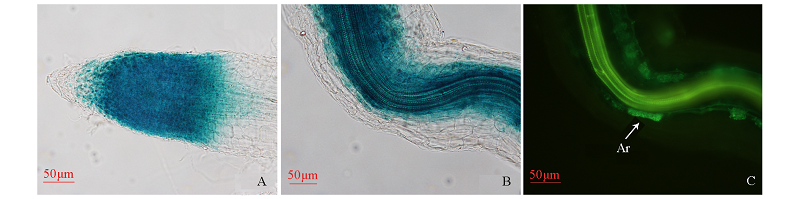
Fig. 7 The GUS histochemical staining of the PtrLYK2 promoter(A,B)and WGA-488 staining of arbuscular mycorrhizal fungi(C) A:proPtrLYK2:GUS was expressed in the root tip cells;B:proPtrLYK2:GUS was expressed in the arbuscules and cells around arbuscules;C:The WGA-488 staining of arbuscules(Ar)of arbuscular mycorrhizal fungi.
| [1] |
Bago B, Pfeffer P E, Shachar-Hill Y. 2000. Carbon metabolism and transport in arbuscular mycorrhizas. Plant Physiology, 124:949-958.
pmid: 11080273 |
| [2] | Bailey T L, Boden M, Buske F A, Frith M, Grant C E, Clementi L, Ren J, Li W W, Noble W S. 2009. MEME SUITE:tools for motif discovery and searching. Nucleic Acids Research, 37:W202-W208. |
| [3] | Bono J J, Fliegmann J, Gough C, Anna P B, Benoit L, Maria A K, Dörte K H, Frank L W T, René G, Julie V C, Theodorus W J G. 2019. Expression and function of the Medicago truncatula lysin motif receptor-like kinase(LysM‐RLK)gene family in the legume-rhizobia symbiosis. The Model Legume Medicago Truncatula, 6 (8):439-447. |
| [4] |
Buendia L, Wang T, Girardin A, Lefebvre B. 2016. The LysMreceptor-like kinase SlLYK10 regulates the arbuscular mycorrhizal symbiosis in tomato. New Phytologist, 210:184-195.
doi: 10.1111/nph.13753 pmid: 26612325 |
| [5] | Chen C, Chen H, He Y, Xia R. 2018. TBtools,a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv, https://doi.org/10.1101/289660. |
| [6] | Cheng Y, Guo W, Yi H, Pang X, Deng X. 2003. An efficient protocol for genomic DNA extraction from Citrus species. Plant Lecular Biology Reporter, 21:177-178. |
| [7] |
Choi J, William S, Uta P. 2018. Mechanisms underlying establishment of arbuscular mycorrhizal symbioses. Annual Review of Phytopathology, 56:135-160.
doi: 10.1146/phyto.2018.56.issue-1 URL |
| [8] |
Combier J P, Billy F D, Gamas P, Niebel A, Rivas S. 2008. Trans-regulation of the expression of the transcription factor MtHAP2-1 by a uORF controls root nodule development. Genes & Development, 22:1549-1559.
doi: 10.1101/gad.461808 URL |
| [9] |
Den C R O, Streng A, De M S, Cao Q Q, Polone E, Liu W, Ammiraju J S S, Kudrna D, Wing R, Untergasser A, Bisseling T, Geurts R. 2011. LysM-type mycorrhizal receptor recruited for rhizobium symbiosis in nonlegume Parasponia. Science, 331 (6019):909-912.
doi: 10.1126/science.1198181 URL |
| [10] | Deng Xiu-xin, Shu Huai-rui, Hao Yu-jin, Xu Qiang, Han Ming-yu, Zhang Shao-ling, Duan Chang-qing, Jiang Quan, Yi Gan-jun, Chen Hou-bin. 2018. Review on the development of fruit tree discipline in the past century. Journal of Agriculture,(8):24-34. (in Chinese) |
| 邓秀新, 束怀瑞, 郝玉金, 徐强, 韩明玉, 张绍铃, 段常青, 姜全, 易干军, 陈厚彬. 2018. 果树学科百年发展回顾. 农学学报,(8):24-34. | |
| [11] | El-Gebal S, Mistry J, Bateman A, Eddy S R, Luciani A, Potter S C, Qureshi M, Richardson L J, Salazar G A, Smart A, Sonnhammer E L L, Hirsh L, Paladin L, Piovesan D, Tosatto S C E, Finn R D. 2019. The Pfam protein families database in 2019. Nucleic Acids Research, 47:D427-D432. |
| [12] | Gao Mei, Xin Jian-kang, Jiang Shan. 2021. Bioinformatics analysis of lysin motif receptor-like kinase gene family in Physcomitrella patens. Guihaia, 41 (6):979-988. (in Chinese) |
| 高梅, 辛健康, 姜山. 2021. 小立碗藓LysM型类受体激酶基因家族生物信息学分析. 广西植物, 41 (6):979-988. | |
| [13] | Ge Shibei, Jiang Xiaochun, Wang Lingyu, Yu Jingquan, Zhou Yanhong. 2020. Recent advances in the role and mechanism of arbuscular mycorrhiza-induced improvement of abiotic stress tolerance in horticultural plants. Acta Horticulturae Sinica, 47 (9):1752-1776. (in Chinese) |
| 葛诗蓓, 姜小春, 王羚羽, 喻景权, 周艳虹. 2020. 园艺植物丛枝菌根抗非生物胁迫的作用机制研究进展. 园艺学报, 47 (9):1752-1776. | |
| [14] |
Indrasumunar A, Wilde J, Hayashi S, Li D, Gresshoff P. 2015. Functional analysis of duplicated Symbiosis Receptor Kinase(SymRK)genes during nodulation and mycorrhizal infection in soybean(Glycine max). Journal of Plant Physiology, 176:157-168.
doi: 10.1016/j.jplph.2015.01.002 pmid: 25617765 |
| [15] |
Ivashuta S, Liu J, Liu J, Lohar D P, Haridas S, Bucciarelli B, Vandenbosch K A, Vance C P, Harrison M J, Gantt J S. 2005. RNA interference identifies a calcium-dependent protein kinase involved in Medicago truncatula root development. Plant Cell, 17:2911-2921.
pmid: 16199614 |
| [16] |
Jiang Y N, Xie Q J, Wang W X, Yang J, Zhang X W, Yu Na, Zhou Y, Wang E T. 2018. Medicago AP2-domain transcription factor WRI5a is a master regulator of lipid biosynthesis and transfer during mycorrhizal symbiosis. Molecular Plant, 11 (11):1344-1359.
doi: S1674-2052(18)30301-0 pmid: 30292683 |
| [17] |
Karimi M, Inzé D, Depicker A. 2002. GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science, 7 (5):193-195.
doi: 10.1016/S1360-1385(02)02251-3 URL |
| [18] |
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X:molecular evolutionary genetics analysis across computing Platforms. Molecular Biology Evolution, 35:1547-1549.
doi: 10.1093/molbev/msy096 URL |
| [19] |
Kutschera U, Briggs W R. 2012. Root phototropism:from dogma to the mechanism of blue light perception. Planta, 235 (3):443-452.
doi: 10.1007/s00425-012-1597-y pmid: 22293854 |
| [20] |
Lian X Y, Zhao X Y, Zhao Q, Wang G L, Li Y Y, Hao Y J. 2021. MdDREB2A in apple is involved in the regulation of multiple abiotic stress responses. Horticultural Plant Journal, 7 (3):197-208.
doi: 10.1016/j.hpj.2021.03.006 URL |
| [21] |
Liao D H, Sun X, Wang N, Song F M, Liang Y. 2018. Tomato-LysM receptor-like kinase SlLYK12 is involved in arbuscular mycorrhizal symbiosis. Front. Plant Science, 9:1004.
doi: 10.3389/fpls.2018.01004 URL |
| [22] |
Limpens E, Ramos J, Franken C, Raz V, Compaan B, Franssen H, Bisseling T, Geurts R. 2004. RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. Journal of Experimental Botany, 55:983-992.
pmid: 15073217 |
| [23] |
Lin K, Limpens E, Zhang Z H, Ivanov S, Saunders D G O, Mu D S, Pang E L, Cao H F, Cha H, Lin T, Zhou Q, Shang Y, Li Y, Sharma T, Velzen R V, Ruijter N D, Aanen D K, Win J, Kamoun S, Bisseling T, Geurts R, Huang S W. 2014. Single nucleus genome sequencing reveals high similarity among nuclei of an endomycorrhizal fungus. PLoS Genetics, 10 (1):e1004078.
doi: 10.1371/journal.pgen.1004078 URL |
| [24] | Liu Run-jin, Chen Ying-long. 2007. Mycorrhizology. Beijing: Science Press. (in Chinese) |
| 刘润进, 陈应龙. 2007. 菌根学. 北京: 科学出版社. | |
| [25] | Liu Yan-jing, Sun Gui-lian, Zhou Qin, Zou Xue-mei, Huang Xiao-long. 2020. Bioinformatics analysis of the FvLysM gene family of strawberry in the forest. Molecular Plant Breeding,https://kns.cnki.net/kcms/detail/46.1068.S.20201209.1055.006.html. (in Chinese) |
| 刘艳晶, 孙贵连, 周琴, 邹雪梅, 黄小龙. 2020. 森林草莓FvLysM基因家族的生物信息学分析. 分子植物育种,https://kns.cnki.net/kcms/detail/46.1068.S.20201209.1055.006.html. | |
| [26] |
Lohmann G V, Shimoda Y, Nielsen M W, Jørgensen F G, Grossmann C, Sandal N, Sørensen K, Thirup S, Madsen L H, Tabata S, Sato S, Stougaard J, Radutoiu S. 2010. Evolution and regulation of the Lotus japonicus LysM receptor gene family. Molecular Plant Microbe Interaction, 23 (4):510-521.
doi: 10.1094/MPMI-23-4-0510 URL |
| [27] |
Miyata K, Hayafune M, Kobae Y, Kaku H, Nishizawa Y, Masuda Y, Shibuya N, Nakagawa T. 2016. Evaluation of the role of the LysM receptor-like kinase,OsLYK2/OsRLK2for AM symbiosis in rice. Plant Cell Physiology, 57:2283-2290.
doi: 10.1093/pcp/pcw144 URL |
| [28] |
Miyata K, Kozaki T, Kouzai Y, Ozawa K, Ishii K, Asamizu E, Okabe Y, Umehara Y, Miyamoto A, Kobae Y, Akiyama K, Kaku H, Nishizawa Y, Shibuya N, Nakagawa T. 2014. The bifunctional plant receptor,OsCERK1,regulates both chitin-triggered immunity and arbuscular mycorrhizal symbiosis in rice. Plant Cell Physiology, 55:1864-1872.
doi: 10.1093/pcp/pcu129 URL |
| [29] |
Pimprikar P, Carbonnel S, Paries M, Katzer K, Klingl V, Bohmer M J, Karl L, Floss D S, Harrison M J, Parniske M, Gutjahr C. 2016. A CCaMK-CYCLOPS-DELLA complex activates transcription of RAM1to regulate arbuscule branching. Current Biology, 26 (8):987-998.
doi: 10.1016/j.cub.2016.01.069 pmid: 27020747 |
| [30] |
Radhakrishnan G V, Keller J, Rich M K, Vernié T, Mbadinga D L M, Vigneron N, Cottret L, Clemente H S, Libourel C, Cheema J, Linde A M, Eklund D M, Cheng S F, Wong G K S, Lagercrantz U, Li F W, Oldroyd G E D, Delaux P M. 2020. An ancestral signalling pathway is conserved in intracellular symbioses-forming plant lineages. Nature Plants, 6 (3):280-289.
doi: 10.1038/s41477-020-0613-7 pmid: 32123350 |
| [31] |
Shu B, Wang P, Xia R X. 2015. Characterisation of the phytase gene in trifoliate orange(Poncirus trifoliata (L.) Raf.)seedlings. Scientia Horticulturae, 194:222-229.
doi: 10.1016/j.scienta.2015.08.028 URL |
| [32] |
Smith S E, Smith F A. 2011. Roles of arbuscular mycorrhizas in plant nutrition and growth:new paradigms fromcellular toecosystem scales. Annual Review of Plant Biology, 62:227-250.
doi: 10.1146/arplant.2011.62.issue-1 URL |
| [33] |
Smith S E, Smith F A, Jakobsen I. 2004. Functional diversity in arbuscular mycorrhizal(AM)symbioses:the contribution of the mycorrhizal P uptake pathway is not correlated with mycorrhizal responsesin growth or total P uptake. New Phytologist, 162:511-524.
doi: 10.1111/nph.2004.162.issue-2 URL |
| [34] |
Sun W J, Ji X L, Song L Q, Wang X F, You C X, Hao Y J. 2021. Functional identification of MdSMXL8.2,the homologous gene of strigolactones pathway repressor protein gene in Malus × domestica. Horticultural Plant Journal, 7 (4):275-285.
doi: 10.1016/j.hpj.2021.01.001 URL |
| [35] |
Wu Q S, He J D, Srivastava A K, Zou Y, Kuča K. 2019. Mycorrhizas enhance drought tolerance of citrus by altering root fatty acid compositions and their saturation levels. Tree Physiology, 39 (7):1149-1158.
doi: 10.1093/treephys/tpz039 URL |
| [36] |
Xiao J, Hu C, Chen Y, Yang B, Hua J. 2014. Effects of low magnesium and an arbuscular mycorrhizal fungus on the growth,magnesium distribution and photosynthesis of two citrus cultivars. Scientia Horticulturae, 177:14-20.
doi: 10.1016/j.scienta.2014.07.016 URL |
| [37] |
Xu Q, Chen L L, Ruan X N, Chen D J, Zhu A D, Chen C L, Bertrand D, Jiao W B, Hao B H, Lyon M P, Chen J J, Gao S, Xing F, Lan H, Chang J W, Ge X H, Lei Y, Hu Q, Miao Y, Wang L, Xiao S X, Biswas M K, Zeng W F, Guo F, Cao H B, Yang X M, Xu X W, Cheng Y J, Xu J, Liu J H, Luo O J H, Tang Z H, Guo W W, Kuang H H, Zhang H Y, Roose M L, Nagarajan N, Deng X X. 2013. The draft genome of sweet orange(Citrus sinensis). Nature Genetics, 45:59-66.
doi: 10.1038/ng.2472 URL |
| [38] |
Yang S, Grønlund M, Jakobsen I, Grotemeyer M S, Rentsch D, Miyao A, Hirochik H, Kumar C S, Sundaresan V, Salamin N, Catausan S, Mattes N, Heuer S, Paszkowskia U. 2012. Nonredundant regulation of rice arbuscular mycorrhizal symbiosis by two members of the PHOSPHATE TRANSPORTER1 gene family. Plant Cell, 24:4236-4251.
doi: 10.1105/tpc.112.104901 URL |
| [39] |
Yoo S D, Cho Y H, Sheen J. 2007. Arabidopsis mesophyll protoplasts:a versatile cell system for transient gene expression analysis. Nature Protocols, 2 (7):1565.
doi: 10.1038/nprot.2007.199 URL |
| [40] | Zeng Li, Li Jianfu, Liu Jianfu, Wang Mingyuan. 2014. Effects of AM fungi inoculation on citrus fruit quality under natural conditions. Journal of Southwest Agricultural Sciences, 27 (5):2101-2105. (in Chinese) |
| 曾理, 李建福, 刘建福, 王明元. 2014. 自然条件下接种AM真菌对柑橘果实品质的影响. 西南农业学报, 27 (5):2101-2105. | |
| [41] |
Zhang W E, Pan X J, Zhao Q, Zhao T. 2021. Plant growth,antioxidative enzyme,and cadmium tolerance responses to cadmium stress in Canna orchioides. Horticultural Plant Journal, 7 (3):256-266.
doi: 10.1016/j.hpj.2021.03.003 URL |
| [42] |
Zhang X, Dong W, Sun J, Feng F, Deng Y, He Z, Oldoyd G E D, Wang E. 2015. The receptor kinase CERK1has dual functions insymbiosis and immunity signaling. Plant Journal, 81:258-267.
doi: 10.1111/tpj.2015.81.issue-2 URL |
| [43] |
Zipfel C, Oldoyd G E D. 2017. Plant signaling in symbiosis and immunity. Nature, 543 (7645):328-336.
doi: 10.1038/nature22009 URL |
| [44] | Zuo Cunwu, Zhang Weina, Mao Juan, Jiang Xuefeng, Ma Zonghuan, Su Jing, Chen Baihong. 2017. Identification and expression of apple LysM receptor kinase genes. Acta Horticulturae Sinica, 44 (4):733-742. (in Chinese) |
| 左存武, 张卫娜, 毛娟, 姜雪峰, 马宗桓, 苏静, 陈佰鸿. 2017. 苹果LysM类受体激酶基因家族鉴定与表达分析. 园艺学报, 44 (4):733-742. |
| [1] | YU Tingting, LI Huan, NING Yuansheng, SONG Jianfei, PENG Lulin, JIA Junqi, ZHANG Weiwei, and YANG Hongqiang. Genome-wide Identification of GRAS Gene Family in Apple and Expression Analysis of Its Response to Auxin [J]. Acta Horticulturae Sinica, 2023, 50(2): 397-409. |
| [2] | YUAN Xin, XU Yunhe, ZHANG Yupei, SHAN Nan, CHEN Chuying, WAN Chunpeng, KAI Wenbin, ZHAI Xiawan, CHEN Jinyin, GAN Zengyu. Studies on AcAREB1 Regulating the Expression of AcGH3.1 During Postharvest Ripening of Kiwifruit [J]. Acta Horticulturae Sinica, 2023, 50(1): 53-64. |
| [3] | WANG Sha, ZHANG Xinhui, ZHAO Yujie, LI Bianbian, ZHAO Xueqing, SHEN Yu, DONG Jianmei, YUAN Zhaohe. Cloning and Functional Analysis of PgMYB111 Related to Anthocyanin Synthesis in Pomegranate [J]. Acta Horticulturae Sinica, 2022, 49(9): 1883-1894. |
| [4] | ZHENG Lin, WANG Shuai, LIU Yunuo, DU Meixia, PENG Aihong, HE Yongrui, CHEN Shanchun, ZOU Xiuping. Gene Cloning and Expression Analysis of NAC Gene in Citrus in Response to Huanglongbing [J]. Acta Horticulturae Sinica, 2022, 49(7): 1441-1457. |
| [5] | WEI Zhuangmin, WEI Sijia, CHEN Peng, HU Jianbing, TANG Yuqing, YE Junli, LI Xianxin, DENG Xiuxin, CHAI Lijun. Identification of S-genotypes of 63 Pummelo Germplasm Resources [J]. Acta Horticulturae Sinica, 2022, 49(5): 1111-1120. |
| [6] | WANG Dan, WANG Mi, LIU Jun, ZHOU Xiaohui, LIU Songyu, YANG Yan, ZHUANG Yong. Cloning of U6 Promoters and Establishment of CRISPR/Cas9 Mediated Gene Editing System in Eggplant [J]. Acta Horticulturae Sinica, 2022, 49(4): 791-800. |
| [7] | WANG Ying, QIN Yangyang, ZENG Ting, LIAO Ping, ZHANG Wei, ZHOU Yan, ZHOU Changyong. Effects of Citrus Yellow Vein Clearing Virus on Photosynthetic Characteristics and Chloroplast Ultrastructure of Lemon [J]. Acta Horticulturae Sinica, 2022, 49(4): 861-867. |
| [8] | XIANG Li, ZHAO Lei, WANG Mei, LÜ Yi, WANG Yanfang, SHEN Xiang, CHEN Xuesen, YIN Chengmiao, MAO Zhiquan. Cloning and Functional Analysis of MdWRKY74 in Apple [J]. Acta Horticulturae Sinica, 2022, 49(3): 482-492. |
| [9] | HUANG Renwei, REN Yinghong, QI Weiliang, ZENG Rui, LIU Xinyu, DENG Binyan. Cloning of Mulberry MaERF105-Like Gene and Its Expression Under Drought Stress [J]. Acta Horticulturae Sinica, 2022, 49(11): 2439-2448. |
| [10] | XIE Siyi, ZHOU Chengzhe, ZHU Chen, ZHAN Dongmei, CHEN Lan, WU Zuchun, LAI Zhongxiong, GUO Yuqiong. Genome-wide Identification and Expression Analysis of CsTIFY Transcription Factor Family Under Abiotic Stress and Hormone Treatments in Camellia sinensis [J]. Acta Horticulturae Sinica, 2022, 49(1): 100-116. |
| [11] | TU Liqin, GAN Shexiang, WU Shuhua, REN Chunmei, CHENG Zhaobang, ZHANG Songbai, ZHU Yuelin, ZHOU Yijun, JI Yinghua. Characterization of the Subcellular Localization and Pathogenicity of P6 Encoded by Cucurbit Chlorotic Yellows Virus [J]. Acta Horticulturae Sinica, 2021, 48(8): 1531-1540. |
| [12] | YANG Tianchen, CHEN Xiaotong, LÜ Ke, ZHANG Di. Expression Pattern and Regulation Mechanism of ApSK3 Dehydrin (Agapanthus praecox)Response to Abiotic Stress and Hormone Signals [J]. Acta Horticulturae Sinica, 2021, 48(8): 1565-1578. |
| [13] | DENG Zeyi, SONG Xiang, HONG Yan, DAI Silan. Applications of Promoters in the Genetic Engineering of Ornamental Plants:A Review [J]. Acta Horticulturae Sinica, 2021, 48(6): 1250-1264. |
| [14] | CAI Roudi, LI Xue, CHEN Yan, XU Xiaoping, CHEN Xiaohui, LAI Zhongxiong, LIN Yuling. Genome-wide Identification and Expression Analysis of DRB Gene Family in Dimocarpus longan [J]. Acta Horticulturae Sinica, 2021, 48(5): 921-933. |
| [15] | LI Zheng, LIU Bing, ZHOU Hong, WANG Xiuyun, XIA Yiping. Isolation and Function Analysis of the Promoter of a Thermal Inducible Gene RCA1 in Rhododendron hainanense [J]. Acta Horticulturae Sinica, 2021, 48(3): 566-576. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd