
Acta Horticulturae Sinica ›› 2022, Vol. 49 ›› Issue (2): 252-264.doi: 10.16420/j.issn.0513-353x.2020-0724
• Research Papers • Previous Articles Next Articles
JIANG Cuicui, FANG Zhizhen, ZHOU Danrong, LIN Yanjuan, YE Xinfu( )
)
Received:2021-05-28
Revised:2021-09-30
Online:2022-02-25
Published:2022-02-28
Contact:
YE Xinfu
E-mail:yexinfu@126.com
CLC Number:
JIANG Cuicui, FANG Zhizhen, ZHOU Danrong, LIN Yanjuan, YE Xinfu. Identification and Expression Analysis of Sugar Transporter Family Genes in‘Furongli’(Prunus salicina)[J]. Acta Horticulturae Sinica, 2022, 49(2): 252-264.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2020-0724
| 序列编号 Gene ID | 基因名称 Gene name | 正向引物(5′-3′) Forward primer | 反向引物(5′-3′) Reverse primer |
|---|---|---|---|
| EVM0028193.1 | PsSUC1 | AAGTCGCCATCGCAGAAGG | AGAGCCAGACGAAAGCGGA |
| EVM0017205.2 | PsTMT1 | GGTATTGTAATGCTGTGGGCTC | CCTTATTTCAGGTGGTGCTGTC |
| EVM0013841.1 | PsPMT6 | CAACCCCAGAAAACCATAGCAGA | CCGAGCCGATGAGGGAGTAAAG |
| EVM0025082.1 | PsINT2 | CCAGATTGCGGGTTTTGTGC | AAAGTGCCAAGCCAGTTAGTGC |
| EVM0027680.1 | PsINT4 | TTACAGCCCAACAATCGTTCAGT | GCCTTCTCCTCCCATATCTATCCA |
| EVM0013528.1 | PsSFP4 | TGGCAACTGGATACGGAATGG | ATGGTCACTGCACAGGGAACG |
| EVM0002384.1 | PsSFP5 | ATGGTCACTGCACAGGGAACG | GCTGACATTCTCATTGCCCCTTT |
| EVM0021731.1 | PsSTP11 | AAGCAGGGCTCCTTGTCCATA | CCAGAGCAGCAAGATACAGCA |
| EVM0014989.1 | PsSTP17 | CTTGCTTGGTGTTGGGGTTGG | GGTGGTCGTTGGTGCTTTGGATA |
Table 1 Primer pairs used in qRT-PCR analysis
| 序列编号 Gene ID | 基因名称 Gene name | 正向引物(5′-3′) Forward primer | 反向引物(5′-3′) Reverse primer |
|---|---|---|---|
| EVM0028193.1 | PsSUC1 | AAGTCGCCATCGCAGAAGG | AGAGCCAGACGAAAGCGGA |
| EVM0017205.2 | PsTMT1 | GGTATTGTAATGCTGTGGGCTC | CCTTATTTCAGGTGGTGCTGTC |
| EVM0013841.1 | PsPMT6 | CAACCCCAGAAAACCATAGCAGA | CCGAGCCGATGAGGGAGTAAAG |
| EVM0025082.1 | PsINT2 | CCAGATTGCGGGTTTTGTGC | AAAGTGCCAAGCCAGTTAGTGC |
| EVM0027680.1 | PsINT4 | TTACAGCCCAACAATCGTTCAGT | GCCTTCTCCTCCCATATCTATCCA |
| EVM0013528.1 | PsSFP4 | TGGCAACTGGATACGGAATGG | ATGGTCACTGCACAGGGAACG |
| EVM0002384.1 | PsSFP5 | ATGGTCACTGCACAGGGAACG | GCTGACATTCTCATTGCCCCTTT |
| EVM0021731.1 | PsSTP11 | AAGCAGGGCTCCTTGTCCATA | CCAGAGCAGCAAGATACAGCA |
| EVM0014989.1 | PsSTP17 | CTTGCTTGGTGTTGGGGTTGG | GGTGGTCGTTGGTGCTTTGGATA |
| 序列编号 Gene ID | 基因名称 Gene name | CDS/bp | 蛋白质/aa Protein | 分子量/kD Molecular weigh | 等电点 pI | 亚细胞定位 Cell location | E值 E-value |
|---|---|---|---|---|---|---|---|
| EVM0028115.1 | PsSTP1 | 1 533 | 511 | 55.32 | 9.53 | 液泡膜 Vacuolar membrane | 7.10E-111 |
| EVM0005102.1 | PsSTP2 | 1 551 | 517 | 56.19 | 9.61 | 液泡膜 Vacuolar membrane | 4.00E-119 |
| EVM0027704.1 | PsSTP3 | 1 596 | 532 | 58.07 | 9.16 | 质膜 Plasma membrane | 5.80E-127 |
| EVM0008700.1 | PsSTP4 | 1 542 | 514 | 55.97 | 8.70 | 质膜 Plasma membrane | 8.60E-128 |
| EVM0000375.1 | PsSTP5 | 1 584 | 528 | 57.87 | 9.24 | 液泡膜 Vacuolar membrane | 1.50E-122 |
| EVM0026713.1 | PsSTP6 | 1 533 | 511 | 55.91 | 9.42 | 液泡膜 Vacuolar membrane | 2.50E-124 |
| EVM0020907.1 | PsSTP7 | 1 533 | 511 | 56.06 | 9.55 | 液泡膜 Vacuolar membrane | 1.50E-122 |
| EVM0019545.1 | PsSTP8 | 1 404 | 468 | 52.71 | 9.76 | 液泡膜 Vacuolar membrane | 5.70E-87 |
| EVM0015455.1 | PsSTP9 | 1 503 | 510 | 56.54 | 8.30 | 质膜 Plasma membrane | 4.60E-125 |
| EVM0003755.1 | PsSTP10 | 1 545 | 515 | 56.77 | 10.07 | 质膜 Plasma membrane | 5.90E-118 |
| EVM0021731.1 | PsSTP11 | 1 509 | 503 | 55.18 | 9.84 | 质膜 Plasma membrane | 1.00E-117 |
| EVM0025512.1 | PsSTP12 | 1 545 | 515 | 55.98 | 9.25 | 质膜 Plasma membrane | 4.30E-119 |
| EVM0006054.1 | PsSTP13 | 1 566 | 522 | 57.67 | 9.14 | 质膜 Plasma membrane | 6.40E-136 |
| EVM0000188.1 | PsSTP14 | 1 551 | 517 | 55.53 | 9.67 | 质膜 Plasma membrane | 1.20E-124 |
| EVM0006545.1 | PsSTP15 | 1 551 | 517 | 55.66 | 9.65 | 质膜 Plasma membrane | 1.70E-125 |
| EVM0015287.1 | PsSTP16 | 1 524 | 508 | 55.39 | 9.88 | 质膜 Plasma membrane | 1.30E-118 |
| EVM0014989.1 | PsSTP17 | 1 317 | 439 | 47.73 | 10.04 | 质膜 Plasma membrane | 2.80E-77 |
| EVM0006896.1 | PsSTP18 | 1 521 | 507 | 55.23 | 8.93 | 液泡膜 Vacuolar membrane | 9.80E-114 |
| EVM0025706.1 | PsSFP1 | 1 467 | 489 | 52.65 | 8.38 | 质膜 Plasma membrane | 7.60E-84 |
| EVM0002206.1 | PsSFP2 | 1 458 | 486 | 52.44 | 9.00 | 质膜 Plasma membrane | 1.30E-98 |
| EVM0026716.1 | PsSFP3 | 939 | 313 | 34.10 | 8.57 | 质膜 Plasma membrane | 2.30E-46 |
| EVM0013528.1 | PsSFP4 | 2 844 | 948 | 102.12 | 8.59 | 质膜 Plasma membrane | 6.30E-169 |
| EVM0002384.1 | PsSFP5 | 1 482 | 494 | 53.81 | 6.65 | 质膜 Plasma membrane | 1.70E-70 |
| EVM0019887.1 | PspGlcT1 | 1 611 | 537 | 58.56 | 8.20 | 质膜 Plasma membrane | 1.00E-99 |
| EVM0017959.1 | PspGlcT2 | 1 500 | 500 | 54.02 | 8.54 | 质膜 Plasma membrane | 1.30E-94 |
| EVM0012278.1 | PspGlcT3 | 1 368 | 456 | 48.44 | 7.68 | 液泡膜 Vacuolar membrane | 3.00E-58 |
| EVM0010156.1 | PspGlcT4 | 1 647 | 549 | 57.35 | 9.62 | 质膜 Plasma membrane | 6.10E-106 |
| EVM0002496.1 | PsINT1 | 1 494 | 498 | 53.86 | 4.79 | 质膜 Plasma membrane | 5.10E-123 |
| EVM0025082.1 | PsINT2 | 1 731 | 577 | 62.94 | 8.80 | 质膜 Plasma membrane | 4.10E-123 |
| EVM0018913.1 | PsINT3 | 1 734 | 578 | 62.65 | 9.00 | 质膜 Plasma membrane | 1.30E-119 |
| EVM0027680.1 | PsINT4 | 1 728 | 576 | 62.02 | 8.62 | 质膜 Plasma membrane | 2.40E-116 |
| EVM0024467.1 | PsPMT1 | 1 584 | 528 | 57.42 | 8.76 | 质膜 Plasma membrane | 6.10E-106 |
| EVM0006809.1 | PsPMT2 | 1 575 | 525 | 56.92 | 9.70 | 质膜 Plasma membrane | 4.80E-106 |
| EVM0013455.1 | PsPMT3 | 1 524 | 508 | 54.47 | 5.98 | 质膜 Plasma membrane | 6.70E-104 |
| EVM0010693.1 | PsPMT4 | 1 587 | 529 | 56.73 | 5.07 | 液泡膜 Vacuolar membrane | 7.10E-110 |
| EVM0014524.1 | PsPMT5 | 1 473 | 491 | 53.14 | 9.63 | 质膜 Plasma membrane | 9.80E-93 |
| EVM0013841.1 | PsPMT6 | 1 614 | 538 | 58.37 | 9.44 | 质膜 Plasma membrane | 2.80E-108 |
| EVM0016266.1 | PsPMT7 | 1 593 | 531 | 57.74 | 8.28 | 质膜 Plasma membrane | 4.50E-104 |
| EVM0014991.1 | PsPMT8 | 1 533 | 511 | 55.69 | 8.96 | 质膜 Plasma membrane | 5.40E-105 |
| EVM0010139.1 | PsPMT9 | 1 527 | 509 | 55.44 | 8.34 | 质膜 Plasma membrane | 9.40E-104 |
| EVM0020369.1 | PsPMT10 | 1 641 | 547 | 58.86 | 7.28 | 质膜 Plasma membrane | 6.10E-107 |
| EVM0007278.1 | PsPMT11 | 1 497 | 499 | 54.02 | 9.59 | 液泡膜 Vacuolar membrane | 9.70E-109 |
| EVM0017205.2 | PsTMT1 | 2 208 | 736 | 78.40 | 5.24 | 质膜 Plasma membrane | 1.80E-92 |
| EVM0000294.1 | PsTMT2 | 2 226 | 742 | 79.99 | 5.09 | 质膜 Plasma membrane | 2.70E-96 |
| EVM0025033.1 | PsTMT3 | 2 178 | 726 | 77.92 | 4.97 | 质膜 Plasma membrane | 4.80E-84 |
| EVM0024253.1 | PsTMT4 | 2 217 | 739 | 79.07 | 4.56 | 质膜 Plasma membrane | 1.10E-90 |
| EVM0028193.1 | PsSUC1 | 1 533 | 511 | 53.90 | 9.07 | 质膜 Plasma membrane | 3.10E-80 |
| EVM0013799.1 | PsSUC2 | 1 827 | 609 | 65.44 | 7.03 | 质膜 Plasma membrane | 4.20E-94 |
| EVM0001814.2 | PsSUC3 | 1 497 | 499 | 53.68 | 9.21 | 质膜 Plasma membrane | 5.10E-92 |
Table 2 Structural and biochemical information of sugar transporter family members in‘Furongli’plum
| 序列编号 Gene ID | 基因名称 Gene name | CDS/bp | 蛋白质/aa Protein | 分子量/kD Molecular weigh | 等电点 pI | 亚细胞定位 Cell location | E值 E-value |
|---|---|---|---|---|---|---|---|
| EVM0028115.1 | PsSTP1 | 1 533 | 511 | 55.32 | 9.53 | 液泡膜 Vacuolar membrane | 7.10E-111 |
| EVM0005102.1 | PsSTP2 | 1 551 | 517 | 56.19 | 9.61 | 液泡膜 Vacuolar membrane | 4.00E-119 |
| EVM0027704.1 | PsSTP3 | 1 596 | 532 | 58.07 | 9.16 | 质膜 Plasma membrane | 5.80E-127 |
| EVM0008700.1 | PsSTP4 | 1 542 | 514 | 55.97 | 8.70 | 质膜 Plasma membrane | 8.60E-128 |
| EVM0000375.1 | PsSTP5 | 1 584 | 528 | 57.87 | 9.24 | 液泡膜 Vacuolar membrane | 1.50E-122 |
| EVM0026713.1 | PsSTP6 | 1 533 | 511 | 55.91 | 9.42 | 液泡膜 Vacuolar membrane | 2.50E-124 |
| EVM0020907.1 | PsSTP7 | 1 533 | 511 | 56.06 | 9.55 | 液泡膜 Vacuolar membrane | 1.50E-122 |
| EVM0019545.1 | PsSTP8 | 1 404 | 468 | 52.71 | 9.76 | 液泡膜 Vacuolar membrane | 5.70E-87 |
| EVM0015455.1 | PsSTP9 | 1 503 | 510 | 56.54 | 8.30 | 质膜 Plasma membrane | 4.60E-125 |
| EVM0003755.1 | PsSTP10 | 1 545 | 515 | 56.77 | 10.07 | 质膜 Plasma membrane | 5.90E-118 |
| EVM0021731.1 | PsSTP11 | 1 509 | 503 | 55.18 | 9.84 | 质膜 Plasma membrane | 1.00E-117 |
| EVM0025512.1 | PsSTP12 | 1 545 | 515 | 55.98 | 9.25 | 质膜 Plasma membrane | 4.30E-119 |
| EVM0006054.1 | PsSTP13 | 1 566 | 522 | 57.67 | 9.14 | 质膜 Plasma membrane | 6.40E-136 |
| EVM0000188.1 | PsSTP14 | 1 551 | 517 | 55.53 | 9.67 | 质膜 Plasma membrane | 1.20E-124 |
| EVM0006545.1 | PsSTP15 | 1 551 | 517 | 55.66 | 9.65 | 质膜 Plasma membrane | 1.70E-125 |
| EVM0015287.1 | PsSTP16 | 1 524 | 508 | 55.39 | 9.88 | 质膜 Plasma membrane | 1.30E-118 |
| EVM0014989.1 | PsSTP17 | 1 317 | 439 | 47.73 | 10.04 | 质膜 Plasma membrane | 2.80E-77 |
| EVM0006896.1 | PsSTP18 | 1 521 | 507 | 55.23 | 8.93 | 液泡膜 Vacuolar membrane | 9.80E-114 |
| EVM0025706.1 | PsSFP1 | 1 467 | 489 | 52.65 | 8.38 | 质膜 Plasma membrane | 7.60E-84 |
| EVM0002206.1 | PsSFP2 | 1 458 | 486 | 52.44 | 9.00 | 质膜 Plasma membrane | 1.30E-98 |
| EVM0026716.1 | PsSFP3 | 939 | 313 | 34.10 | 8.57 | 质膜 Plasma membrane | 2.30E-46 |
| EVM0013528.1 | PsSFP4 | 2 844 | 948 | 102.12 | 8.59 | 质膜 Plasma membrane | 6.30E-169 |
| EVM0002384.1 | PsSFP5 | 1 482 | 494 | 53.81 | 6.65 | 质膜 Plasma membrane | 1.70E-70 |
| EVM0019887.1 | PspGlcT1 | 1 611 | 537 | 58.56 | 8.20 | 质膜 Plasma membrane | 1.00E-99 |
| EVM0017959.1 | PspGlcT2 | 1 500 | 500 | 54.02 | 8.54 | 质膜 Plasma membrane | 1.30E-94 |
| EVM0012278.1 | PspGlcT3 | 1 368 | 456 | 48.44 | 7.68 | 液泡膜 Vacuolar membrane | 3.00E-58 |
| EVM0010156.1 | PspGlcT4 | 1 647 | 549 | 57.35 | 9.62 | 质膜 Plasma membrane | 6.10E-106 |
| EVM0002496.1 | PsINT1 | 1 494 | 498 | 53.86 | 4.79 | 质膜 Plasma membrane | 5.10E-123 |
| EVM0025082.1 | PsINT2 | 1 731 | 577 | 62.94 | 8.80 | 质膜 Plasma membrane | 4.10E-123 |
| EVM0018913.1 | PsINT3 | 1 734 | 578 | 62.65 | 9.00 | 质膜 Plasma membrane | 1.30E-119 |
| EVM0027680.1 | PsINT4 | 1 728 | 576 | 62.02 | 8.62 | 质膜 Plasma membrane | 2.40E-116 |
| EVM0024467.1 | PsPMT1 | 1 584 | 528 | 57.42 | 8.76 | 质膜 Plasma membrane | 6.10E-106 |
| EVM0006809.1 | PsPMT2 | 1 575 | 525 | 56.92 | 9.70 | 质膜 Plasma membrane | 4.80E-106 |
| EVM0013455.1 | PsPMT3 | 1 524 | 508 | 54.47 | 5.98 | 质膜 Plasma membrane | 6.70E-104 |
| EVM0010693.1 | PsPMT4 | 1 587 | 529 | 56.73 | 5.07 | 液泡膜 Vacuolar membrane | 7.10E-110 |
| EVM0014524.1 | PsPMT5 | 1 473 | 491 | 53.14 | 9.63 | 质膜 Plasma membrane | 9.80E-93 |
| EVM0013841.1 | PsPMT6 | 1 614 | 538 | 58.37 | 9.44 | 质膜 Plasma membrane | 2.80E-108 |
| EVM0016266.1 | PsPMT7 | 1 593 | 531 | 57.74 | 8.28 | 质膜 Plasma membrane | 4.50E-104 |
| EVM0014991.1 | PsPMT8 | 1 533 | 511 | 55.69 | 8.96 | 质膜 Plasma membrane | 5.40E-105 |
| EVM0010139.1 | PsPMT9 | 1 527 | 509 | 55.44 | 8.34 | 质膜 Plasma membrane | 9.40E-104 |
| EVM0020369.1 | PsPMT10 | 1 641 | 547 | 58.86 | 7.28 | 质膜 Plasma membrane | 6.10E-107 |
| EVM0007278.1 | PsPMT11 | 1 497 | 499 | 54.02 | 9.59 | 液泡膜 Vacuolar membrane | 9.70E-109 |
| EVM0017205.2 | PsTMT1 | 2 208 | 736 | 78.40 | 5.24 | 质膜 Plasma membrane | 1.80E-92 |
| EVM0000294.1 | PsTMT2 | 2 226 | 742 | 79.99 | 5.09 | 质膜 Plasma membrane | 2.70E-96 |
| EVM0025033.1 | PsTMT3 | 2 178 | 726 | 77.92 | 4.97 | 质膜 Plasma membrane | 4.80E-84 |
| EVM0024253.1 | PsTMT4 | 2 217 | 739 | 79.07 | 4.56 | 质膜 Plasma membrane | 1.10E-90 |
| EVM0028193.1 | PsSUC1 | 1 533 | 511 | 53.90 | 9.07 | 质膜 Plasma membrane | 3.10E-80 |
| EVM0013799.1 | PsSUC2 | 1 827 | 609 | 65.44 | 7.03 | 质膜 Plasma membrane | 4.20E-94 |
| EVM0001814.2 | PsSUC3 | 1 497 | 499 | 53.68 | 9.21 | 质膜 Plasma membrane | 5.10E-92 |
| 糖 Sugar | PsSUC1 | PsTMT1 | PsTMT4 | PsPMT2 | PsPMT4 | PsPMT6 | PsINT2 | PsINT4 |
|---|---|---|---|---|---|---|---|---|
| 葡萄糖 Glucose | -0.93 | 0.74 | 0.66 | 0.11 | 0.51 | -0.95 | -0.87 | -0.87 |
| 果糖 Fructose | 0.61 | -0.21 | -0.23 | -0.01 | -0.31 | 0.65 | 0.24 | 0.52 |
| 蔗糖 Sucrose | 0.83 | -0.81 | -0.61 | -0.13 | -0.63 | 0.93 | 0.82 | 0.94 |
| 糖 Sugar | PsSFP4 | PsSFP5 | PsSTP1 | PsSTP11 | PsSTP12 | PsSTP16 | PsSTP17 | |
| 葡萄糖 Glucose | 0.81 | 0.80 | 0.18 | 0.84 | 0.52 | -0.31 | -0.81 | |
| 果糖 Fructose | -0.57 | -0.56 | 0.12 | -0.73 | -0.58 | 0.49 | 0.64 | |
| 蔗糖 Sucrose | -0.85 | -0.87 | -0.25 | -0.83 | -0.52 | 0.26 | 0.84 |
Table 3 Correlation evaluation between sugars in‘Furongli’fruit during development and PsSTs expression
| 糖 Sugar | PsSUC1 | PsTMT1 | PsTMT4 | PsPMT2 | PsPMT4 | PsPMT6 | PsINT2 | PsINT4 |
|---|---|---|---|---|---|---|---|---|
| 葡萄糖 Glucose | -0.93 | 0.74 | 0.66 | 0.11 | 0.51 | -0.95 | -0.87 | -0.87 |
| 果糖 Fructose | 0.61 | -0.21 | -0.23 | -0.01 | -0.31 | 0.65 | 0.24 | 0.52 |
| 蔗糖 Sucrose | 0.83 | -0.81 | -0.61 | -0.13 | -0.63 | 0.93 | 0.82 | 0.94 |
| 糖 Sugar | PsSFP4 | PsSFP5 | PsSTP1 | PsSTP11 | PsSTP12 | PsSTP16 | PsSTP17 | |
| 葡萄糖 Glucose | 0.81 | 0.80 | 0.18 | 0.84 | 0.52 | -0.31 | -0.81 | |
| 果糖 Fructose | -0.57 | -0.56 | 0.12 | -0.73 | -0.58 | 0.49 | 0.64 | |
| 蔗糖 Sucrose | -0.85 | -0.87 | -0.25 | -0.83 | -0.52 | 0.26 | 0.84 |
| 品种 Cultivar | 葡萄糖 Glucose | 果糖 Fructose | 蔗糖 Sucrose | 总糖 Total soluble carbohydrates |
|---|---|---|---|---|
| 芙蓉李 Furongli | 50.95 ± 3.07 a | 37.89 ± 3.68 a | 22.98 ± 3.95 ab | 111.82 ±10.7 a |
| 秋姬李 Qiujili | 48.08 ± 3.62 a | 38.34 ± 2.26 a | 15.11 ± 2.09 ab | 101.53 ±7.97 ab |
| 红李 Hongli | 39.65 ± 3.12 ab | 33.25 ± 3.84 ab | 4.02 ± 2.25 b | 76.92 ±9.21 b |
| 胭脂李 Yanzhili | 26.71 ± 2.62 b | 26.76 ± 3.20 ab | 25.92 ± 2.73 ab | 79.39 ±8.55 b |
| 华蜜大蜜李 Huami Damili | 25.57 ± 3.27 b | 25.73 ± 3.29 ab | 29.31 ± 4.17 a | 80.61 ±10.73 b |
| 白脆鸡麻李 Baicui Jimali | 23.31 ± 3.07 b | 16.83 ± 2.97 b | 22.99 ± 3.07 ab | 63.13 ±9.11 b |
| 红线李 Hongxianli | 20.90 ± 2.81 b | 24.79 ± 3.05 ab | 15.53 ± 1.15 ab | 61.22 ±7.01 b |
| 黑琥珀 Heihupo | 21.84 ± 2.15 b | 22.97 ± 3.15 b | 5.84 ± 0.51 b | 50.65 ±5.81 b |
Table 4 Analysis of soluble carbohydrates contents in different plum varieties mg · g-1 FW
| 品种 Cultivar | 葡萄糖 Glucose | 果糖 Fructose | 蔗糖 Sucrose | 总糖 Total soluble carbohydrates |
|---|---|---|---|---|
| 芙蓉李 Furongli | 50.95 ± 3.07 a | 37.89 ± 3.68 a | 22.98 ± 3.95 ab | 111.82 ±10.7 a |
| 秋姬李 Qiujili | 48.08 ± 3.62 a | 38.34 ± 2.26 a | 15.11 ± 2.09 ab | 101.53 ±7.97 ab |
| 红李 Hongli | 39.65 ± 3.12 ab | 33.25 ± 3.84 ab | 4.02 ± 2.25 b | 76.92 ±9.21 b |
| 胭脂李 Yanzhili | 26.71 ± 2.62 b | 26.76 ± 3.20 ab | 25.92 ± 2.73 ab | 79.39 ±8.55 b |
| 华蜜大蜜李 Huami Damili | 25.57 ± 3.27 b | 25.73 ± 3.29 ab | 29.31 ± 4.17 a | 80.61 ±10.73 b |
| 白脆鸡麻李 Baicui Jimali | 23.31 ± 3.07 b | 16.83 ± 2.97 b | 22.99 ± 3.07 ab | 63.13 ±9.11 b |
| 红线李 Hongxianli | 20.90 ± 2.81 b | 24.79 ± 3.05 ab | 15.53 ± 1.15 ab | 61.22 ±7.01 b |
| 黑琥珀 Heihupo | 21.84 ± 2.15 b | 22.97 ± 3.15 b | 5.84 ± 0.51 b | 50.65 ±5.81 b |
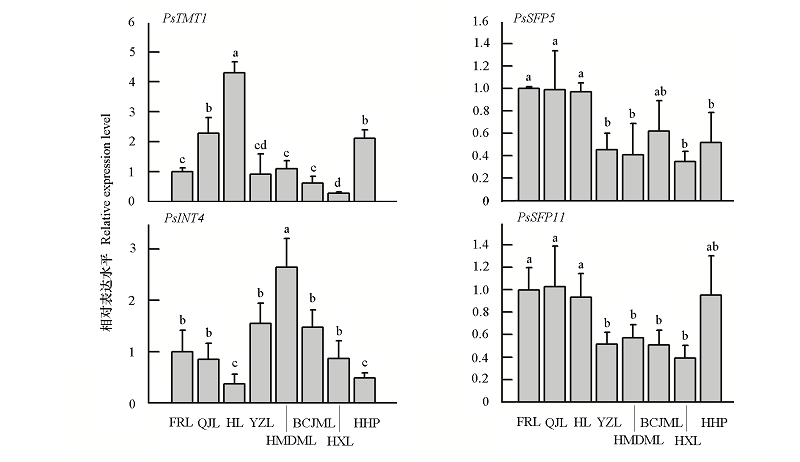
Fig. 6 Quantitative RT-PCR analysis of four sugar transporter genes in different plum cultivar FRL:Furongli;QJL:Qiujili;HL:Hongli;YZL:Yanzhili;HMDML:Huami Damili;BCJML:Baicui Jimali;HXL:Hongxianli;HHP:Heihupo.
| [1] |
Afoufa-Bastien D, Medici A, Jeauffre J, Coutos-Thevenot P, Lemoine R, Atanassova R, Laloi M. 2010. The Vitis vinifera sugar transporter gene family:Phylogenetic overview and microarray expression profiling. BMC Plant Biology, 10:245.
doi: 10.1186/1471-2229-10-245 pmid: 21073695 |
| [2] |
Aluri S, Büttner M. 2007. Identification and functional expression of the Arabidopsis thaliana vacuolar glucose transporter 1 and its role in seed germination and flowering. PNAS, 104 (7):2537-2542.
doi: 10.1073/pnas.0610278104 URL |
| [3] |
Aoki N, Hirose T, Scofield G N, Whitfeld P R, Furbank R T. 2003. The sucrose transporter gene family in rice. Plant and Cell Physiology, 44 (3):223-232.
doi: 10.1093/pcp/pcg030 URL |
| [4] |
Baker R F, Leach K A, Braun D M. 2012. SWEET as sugar:new sucrose effluxers in plants. Molecular Plant, 5 (4):766-768.
doi: 10.1093/mp/SSS054 URL |
| [5] |
Büttner M. 2007. The monosaccharide transporter(-like)gene family in Arabidopsis. FEBS Letters, 581 (12):2318-2324.
doi: 10.1016/j.febslet.2007.03.016 URL |
| [6] | Büttner M. 2010. The Arabidopsis sugar transporter(AtSTP)family:an update. Plant Biology, 12 (s1):35-41. |
| [7] |
Chandran D. 2015. Co-option of developmentally regulated plant SWEET transporters for pathogen nutrition and abiotic stress tolerance. IUBMB Life, 67 (7):461-471.
doi: 10.1002/iub.1394 pmid: 26179993 |
| [8] |
Chen L Q. 2014. SWEET sugar transporters for phloem transport and pathogen nutrition. New Phytologist, 201 (4):1150-1155.
doi: 10.1111/nph.2014.201.issue-4 URL |
| [9] |
Chen L Q, Hou B H, Lalonde S, Takanaga H, Hartung M L, Qu X Q, Guo W J, Kim J G, Underwood W, Chaudhuri B. 2010. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature, 468:527-532.
doi: 10.1038/nature09606 URL |
| [10] |
Cheng R, Cheng Y S, Lu J H, Chen J Q, Wang Y Z, Zhang S L, Zhang H P. 2018. The gene PbTMT4 from pear(Pyrus bretschneideri)mediates vacuolar sugar transport and strongly affects sugar accumulation in fruit. Physiologia Plantarum, 164:307-319.
doi: 10.1111/ppl.2018.164.issue-3 URL |
| [11] |
Cheng R, Zhang H P, Cheng Y S, Wang Y Z, Wang G M, Zhang S L. 2017. In silico and expression analysis of the tonoplast monosaccharide transporter(TMT)gene family in Pyrus bretschneideri. The Journal of Horticultural Science and Biotechnology, 93 (4):366-376.
doi: 10.1080/14620316.2017.1373603 URL |
| [12] |
Decourteix M, Alves G, Bonhomme M, Peuch M, Ben Baazi K, Brunel N, Guilliot A, Rageau R, Ameglio T, Petel G. 2008. Sucrose(JrSUT1)and hexose(JrHT1 and JrHT2)transporters in walnut xylem parenchyma cells:their potential role in early events of growth resumption. Tree Physiology, 28 (2):215-224.
pmid: 18055432 |
| [13] |
Doidy J, Grace E, Kuhn C, Simonplas F, Casieri L, Wipf D. 2012. Sugar transporters in plants and in their interactions with fungi. Trends in Plant Science, 17 (7):413-422.
doi: 10.1016/j.tplants.2012.03.009 pmid: 22513109 |
| [14] |
Gao Z L, Maurousset R, Lemoine S D, Yoo S, Van Nocker S, Loescher W. 2003. Cloning,expression,and characterization of sorbitol transporters from developing sour cherry fruit and leaf sink tissues. Plant Physiology, 131 (4):1566-1575.
doi: 10.1104/pp.102.016725 URL |
| [15] | Geng Yanqiu, Dong Xiaochang, Zhang Chunmei. 2021. Recent progress of sugar transporter in horticultural crops. Acta Horticulturae Sinica, 48 (4):676-688. (in Chinese) |
| 耿艳秋, 董肖昌, 张春梅. 2021. 园艺作物糖转运蛋白研究进展. 园艺学报, 48 (4):676-688. | |
| [16] |
Gong X, Liu M L, Zhang L J, Ruan Y, Ding R, Ji Y Q, Zhang N, Zhang S B, Farmer J, Wang C. 2015. Arabidopsis AtSUC2 and AtSUC4,encoding sucrose transporters,are required for abiotic stress tolerance in an ABA-dependent pathway. Physiologia Plantarum, 153 (1):119-136.
doi: 10.1111/ppl.2015.153.issue-1 URL |
| [17] |
Hayes M A, Dry I B. 2007. Isolation,functional characterization,and expression analysis of grapevine(Vitis vinifera L.)hexose transporters:differential roles in sink and source tissues. Journal of Experimental Botany, 58 (8):1985-1997.
doi: 10.1093/jxb/erm061 URL |
| [18] |
Hu Y B, Sosso D, Qu X Q, Chen L Q, Ma L, Chermak D, Zhang D C, Frommer W B. 2016. Phylogenetic evidence for fusion of archaeal and bacterial SemiSWEETs to form eukaryotic SWEETs and identification of SWEET hexose transporters in the amphibian chytrid pathogen Batrachochytrium dendrobatidis. FASEB Journal, 30 (10):3644-3654.
doi: 10.1096/fsb2.v30.10 URL |
| [19] |
Jiang C C, Fang Z Z, Zhou D R, Pan S L, Ye X F. 2019. Changes in secondary metabolites,organic acids and soluble sugars during the development of plum fruit cv.‘Furongli’(Prunus salicina Lindl.). Journal of the Science of Food and Agriculture, 99:1010-1019.
doi: 10.1002/jsfa.2019.99.issue-3 URL |
| [20] | Jiu S T, Haider M S, Kurjogi M M, Zhang K K, Zhu X D, Fang J G. 2018. Genome-wide characterization and expression analysis of sugar transporter family genes in woodland strawberry. The Plant Genome, 11 (3):1-16. |
| [21] |
Johnson D A, Hill J P, Thomas M A. 2006. The monosaccharide transporter gene family in land plants is ancient and shows differential subfamily expression and expansion across lineages. BMC Evolutionary Biology, 6:64.
doi: 10.1186/1471-2148-6-64 URL |
| [22] |
Johnson D A, Thomas M A. 2007. The monosaccharide transporter gene family in Arabidopsis and rice:a history of duplications,adaptive evolution,and functional divergence. Molecular Biology and Evolution, 24 (11):2412-2423.
doi: 10.1093/molbev/msm184 URL |
| [23] |
Li J M, Zheng D M, Li L T, Qiao X, Wei S W, Bai B, Zhang S L, Wu J. 2015. Genome-wide function,evolutionary characterization and expression analysis of sugar transporter family genes in pear(Pyrus bretschneideri Rehd). Plant and Cell Physiology, 56 (9):1721-1737.
doi: 10.1093/pcp/pcv090 URL |
| [24] | Li Xiao-chuan, Zhou Ping, Wang Chao-hai. 2017. Genome wide identification and expression analysis of sugar transporters in potato. Jiangsu Agricultural Sciences, 45 (12):24-27. (in Chinese) |
| 李晓川, 周平, 王朝海. 2017. 马铃薯糖转运蛋白家族的全基因组鉴定和表达分析. 江苏农业科学, 45 (12):24-27. | |
| [25] |
Liu H T, Ji Y, Liu Y, Tian S H, Gao Q H, Zou X H, Yang J, Dong C, Tan J H, Ni D A, Duan K. 2020. The sugar transporter system of strawberry:genome-wide identification and expression correlation with fruit soluble sugar-related traits in a Fragaria × ananassa germplasm collection. Horticulture Research, 7:132.
doi: 10.1038/s41438-020-00359-0 URL |
| [26] | Liu Shuo, Liu You-chun, Liu Ning, Zhang Yu-ping, Zhang Qiu-ping, Xu Ming, Zhang Yu-jun, Liu Weisheng. 2016. Sugar and organic acid components in fruits of plum cultivar resources of genus Prunus. Scientia Agricultura Sinica, 49 (16):3188-3198. (in Chinese) |
| 刘硕, 刘有春, 刘宁, 张玉萍, 章秋平, 徐铭, 张玉君, 刘威生. 2016. 李属(Prunus)果树品种资源果实糖和酸的组分及其构成差异. 中国农业科学, 49 (16):3188-3198. | |
| [27] |
Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real-time quantitative PCR the 2-ΔΔCT method. Methods, 25,402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [28] | Lu Jing, Ma Qijun, Kang Hui, Li Wenhao, Liu Yajing, Hao Yujing, You Chunxiang. 2019. Ectopic expressing MdSWEET1 in tomato enhanced salt tolerance. Acta Horticulturae Sinica, 46 (3):433-443. (in Chinese) |
| 路静, 马齐军, 康慧, 李文浩, 刘亚静, 郝玉金, 由春香. 2019. 苹果糖转运蛋白基因MdSWEET1在番茄中异源表达提高其耐盐性. 园艺学报, 46 (3):433-443. | |
| [29] |
Reuscher S, Akiyama M, Yasuda T, Makino H, Aoki K, Shibata D, Shiratake K. 2014. The sugar transporter inventory of tomato:genome-wide identification and expression analysis. Plant and Cell Physiology, 55 (6):1123-1141.
doi: 10.1093/pcp/pcu052 pmid: 24833026 |
| [30] |
Schneider S, Schneidereit A, Konrad K R, Hajirezaei M R, Gramann M, Hedrich R, Hedrich R, Sauer N. 2006. Arabidopsis INOSITOL TRANSPORTER4 mediates high-affifinity H+ symport of myoinositol across the plasma membrane. Plant Physiology, 141:565-577.
doi: 10.1104/pp.106.077123 pmid: 16603666 |
| [31] |
Schneider S, Schneidereit A, Udvardi P, Hammes U, Gramann M, Dietrich P, Sauer N. 2007. Arabidopsis INOSITOL TRANSPORTER 2 mediates H+ symport of different inositol epimers and derivatives across the plasma membrane. Plant Physiology, 145:1395-1407.
doi: 10.1104/pp.107.109033 pmid: 17951450 |
| [32] | Shiratake K. 2007. Genetics of sucrose transporters in plants. Genes Genomes Genomics, 1:73-80. |
| [33] |
Slewinski T L. 2011. Diverse functional roles of monosaccharide transporters and their homologs in vascular plants:a physiological perspective. Molecular Plant, 4 (4):641-662.
doi: 10.1093/mp/ssr051 pmid: 21746702 |
| [34] |
Watari J, Kobae Y, Yamaki S, Yamada K, Toyofuku K, Tabuchi T, Shiratake K. 2004. Identification of sorbitol transporters expressed in the phloem of apple source leaves. Plant and Cell Physiology, 45 (8):1032-1041.
doi: 10.1093/pcp/pch121 pmid: 15356329 |
| [35] |
Williams L E, Lemoine R, Sauer N. 2000. Sugar transporters in higher plants -a diversity of roles and complex regulation. Trends Plant Science, 5 (7):283-290.
doi: 10.1016/S1360-1385(00)01681-2 URL |
| [36] | Xuan Y H, Hu Y B, Chen L Q, Sosso D, Ducat D C, Hou B H, Frommer W B. 2013. Functional role of oligomerization for bacterial and plant SWEET sugar transporter family. PNAS, 110 (39):3685-3694. |
| [1] | WANG Rui, HONG Wenjuan, LUO Hua, ZHAO Lina, CHEN Ying, and WANG Jun, . Construction of SSR Fingerprints of Pomegranate Cultivars and Male Parent Identification of Hybrids [J]. Acta Horticulturae Sinica, 2023, 50(2): 265-278. |
| [2] | REN Hailong, XU Donglin, ZHANG Jing, ZOU Jiwen, LI Guangguang, ZHOU Xianyu, XIAO Wanyu, and SUN Yijia. Establishment of SNP Fingerprinting and Identification of Chinese Flowering Cabbage Varieties Based on KASP Genotyping [J]. Acta Horticulturae Sinica, 2023, 50(2): 307-318. |
| [3] | YU Tingting, LI Huan, NING Yuansheng, SONG Jianfei, PENG Lulin, JIA Junqi, ZHANG Weiwei, and YANG Hongqiang. Genome-wide Identification of GRAS Gene Family in Apple and Expression Analysis of Its Response to Auxin [J]. Acta Horticulturae Sinica, 2023, 50(2): 397-409. |
| [4] | LIANG Jiali, WU Qisong, CHEN Guangquan, ZHANG Rong, XU Chunxiang, and FENG Shujie, . Identification of the Neopestalotiopsis musae Pathogen of Banana Leaf Spot Disease [J]. Acta Horticulturae Sinica, 2023, 50(2): 410-420. |
| [5] | ZHAI Hanhan, ZHAI Yujie, TIAN Yi, ZHANG Ye, YANG Li, WEN Zhiliang, CHEN Haijiang. Genome-wide Identification of Peach SAUR Gene Family and Characterization of PpSAUR5 Gene [J]. Acta Horticulturae Sinica, 2023, 50(1): 1-14. |
| [6] | JIANG Jingdong, WEI Zhuangmin, WANG Nan, ZHU Chenqiao, YE Junli, XIE Zongzhou, DENG Xiuxin, CHAI Lijun. Exploitation and Identification of Tetraploid Resources of Hongkong Kumquat(Fortunella hindsii) [J]. Acta Horticulturae Sinica, 2023, 50(1): 27-35. |
| [7] | CUI Jian, ZHONG Xionghui, LIU Zeci, CHEN Denghui, LI Hailong, HAN Rui, YUE Xiangqing, KANG Jungen, WANG Chao. Construction of Cabbage Chromosome Segment Substitution Lines [J]. Acta Horticulturae Sinica, 2023, 50(1): 65-78. |
| [8] | HE Chengyong, ZHAO Xiaoli, XU Tengfei, GAO Dehang, LI Shifang, WANG Hongqing. Genome Sequence Analysis of Strawberry Virus 1 from Shandong Province,China [J]. Acta Horticulturae Sinica, 2023, 50(1): 153-160. |
| [9] | LUO Hailin, YUAN Lei, WENG Hua, YAN Jiahui, GUO Qingyun, WANG Wenqing, MA Xinming. Identification and Analysis of Complete Genomic Sequence of Broad Bean Wilt Virus 2 Pepper Isolate in Qinghai Province [J]. Acta Horticulturae Sinica, 2023, 50(1): 161-169. |
| [10] | ZHAO Xueyan, WANG Qi, WANG Li, WANG Fangyuan, WANG Qing, LI Yan. Comparative Transcriptome Analysis of Differential Expression in Different Tissues of Corydalis yanhusuo [J]. Acta Horticulturae Sinica, 2023, 50(1): 177-187. |
| [11] | DING Zhijie, BAO Jinbo, ROUXIAN Guli, ZHU Tiantian, LI Xueli, MIAO Haoyu, TIAN Xinmin. Comparative Chloroplast Genome Study of Mallus servisii‘Red Delicious’and‘Golden Delicious’ [J]. Acta Horticulturae Sinica, 2022, 49(9): 1977-1990. |
| [12] | YANG Xingyu, XU Linbing, WU Yuanli, QIU Diyang, FAN Linlin, HUANG Bingzhi. Genome(ABBB)Identification of a Hybrid Banana Cultivar‘Fenza 1’ [J]. Acta Horticulturae Sinica, 2022, 49(9): 1991-1997. |
| [13] | GAO Yanlong, WU Yuxia, ZHANG Zhongxing, WANG Shuangcheng, ZHANG Rui, ZHANG De, WANG Yanxiu. Bioinformatics Analysis of Apple ELO Gene Family and Its Expression Analysis Under Low Temperature Stress [J]. Acta Horticulturae Sinica, 2022, 49(8): 1621-1636. |
| [14] | LIU Jinming, GUO Caihua, YUAN Xing, KANG Chao, QUAN Shaowen, NIU Jianxin. Genome-wide Identification of Dof Family Genes and Expression Analysis Sepal Persistent and Abscission in Pear [J]. Acta Horticulturae Sinica, 2022, 49(8): 1637-1649. |
| [15] | QIU Ziwen, LIU Linmin, LIN Yongsheng, LIN Xiaojie, LI Yongyu, WU Shaohua, YANG Chao. Cloning and Functional Analysis of the MbEGS Gene from Melaleuca bracteata [J]. Acta Horticulturae Sinica, 2022, 49(8): 1747-1760. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd