
Acta Horticulturae Sinica ›› 2021, Vol. 48 ›› Issue (12): 2481-2496.doi: 10.16420/j.issn.0513-353x.2020-0717
Previous Articles Next Articles
JIANG Mengqi1, XUE Xiaodong1, SU Liyao1, CHEN Yan1, ZHANG Shuting1, LI Xiaofei1, WANG Peiyu2, ZHANG Zihao1, LAI Zhongxiong1, LIN Yuling1,*( )
)
Received:2021-03-11
Revised:2021-07-30
Published:2022-01-04
Contact:
LIN Yuling
E-mail:buliang84@163.com
CLC Number:
JIANG Mengqi, XUE Xiaodong, SU Liyao, CHEN Yan, ZHANG Shuting, LI Xiaofei, WANG Peiyu, ZHANG Zihao, LAI Zhongxiong, LIN Yuling. Genome-wide Identification and Expression Analysis of TCP Family in Dimocarpus longan[J]. Acta Horticulturae Sinica, 2021, 48(12): 2481-2496.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2020-0717
| 基因名称 Gene name | 基因ID Gene ID | 上游引物(5′-3′) Forward primer | 下游引物(5′-3′) Reverse primer |
|---|---|---|---|
| DlTCP1 | Dlo030081 | TGATCTCCAGGACATGCTAGG | TGCACATCTTTTCTCTGGTCC |
| DlTCP2 | Dlo021979 | CTGTGGAGTTGGATGGTGAAG | GACACTACCAATACCACCAGTCAG |
| DlTCP4a | Dlo009059 | CAATGGGTGCTTCAAACGAC | CCAGCATCATAACCAAGTGGAG |
| DlTCP4b | Dlo027391 | CGTGGCACCCGAATATTAGT | AGTTGATTGATGAGGTCGTGG |
| DlTCP5b | Dlo031128 | CTCATGATCCGAACACCTCTC | CCAGTAGCTAACCATTCTTCCTTAC |
| DlTCP8 | Dlo024507 | TCAGGTGAAGCGGTCGAC | CATAGGGACCCGGATGATG |
| DlTCP9 | Dlo012848 | CGGAGCCAGCTATTATTGC | GTTGTGGCACCATGAAGAAC |
| DlTCP11 | Dlo029447 | CTAACTCACTCCTCCCCGTTAC | ACCCTACAAGACACCGACG |
| DlTCP13 | Dlo030148 | CATCATTCAATGTCATCTTCT | TCAATCCATGGCTGCCTAAT |
| DlTCP15 | Dlo018528 | TCCATAACCGGCGATGAC | GCTCGACCCTGAACTCCTTA |
| DlTCP17 | Dlo026163 | AGAACGCCGTGTCACCTT | TGTTACTCATTCCAGGCGG |
| DlTCP18 | Dlo031253 | ACATGGCCGATTCAACAAG | AACACTCACGCAGCTGCT |
| DlTCP19 | Dlo019709 | GCACCGATCACACCAATGT | TGCTACTCGAACTGGAACTGG |
| DlTCP20a | Dlo019007 | GCTGAGCCATCTATAATTGCTG | CACTCCCATAACTTGATGCC |
| DlTCP20b | Dlo027551 | GAAGCCAGCAGAGATCAAAGAT | GGTATGGTTCCACTCCCTGT |
| DlTCP21 | Dlo017839 | GAGAATCCGGATGCCAAT | TTGTGATCCAGTGAAGTCGAG |
| DlTCP23 | Dlo014979 | CCCACCACGAGAACAGTG | GTAAGGAGACGTTAAGGGAGG |
| Fe-SOD | GGTCAGATGGTGAAGCCGTAGAG | GTCTATGCCACCGATACAACAAACCC |
Table 1 Primers used for qRT-PCR
| 基因名称 Gene name | 基因ID Gene ID | 上游引物(5′-3′) Forward primer | 下游引物(5′-3′) Reverse primer |
|---|---|---|---|
| DlTCP1 | Dlo030081 | TGATCTCCAGGACATGCTAGG | TGCACATCTTTTCTCTGGTCC |
| DlTCP2 | Dlo021979 | CTGTGGAGTTGGATGGTGAAG | GACACTACCAATACCACCAGTCAG |
| DlTCP4a | Dlo009059 | CAATGGGTGCTTCAAACGAC | CCAGCATCATAACCAAGTGGAG |
| DlTCP4b | Dlo027391 | CGTGGCACCCGAATATTAGT | AGTTGATTGATGAGGTCGTGG |
| DlTCP5b | Dlo031128 | CTCATGATCCGAACACCTCTC | CCAGTAGCTAACCATTCTTCCTTAC |
| DlTCP8 | Dlo024507 | TCAGGTGAAGCGGTCGAC | CATAGGGACCCGGATGATG |
| DlTCP9 | Dlo012848 | CGGAGCCAGCTATTATTGC | GTTGTGGCACCATGAAGAAC |
| DlTCP11 | Dlo029447 | CTAACTCACTCCTCCCCGTTAC | ACCCTACAAGACACCGACG |
| DlTCP13 | Dlo030148 | CATCATTCAATGTCATCTTCT | TCAATCCATGGCTGCCTAAT |
| DlTCP15 | Dlo018528 | TCCATAACCGGCGATGAC | GCTCGACCCTGAACTCCTTA |
| DlTCP17 | Dlo026163 | AGAACGCCGTGTCACCTT | TGTTACTCATTCCAGGCGG |
| DlTCP18 | Dlo031253 | ACATGGCCGATTCAACAAG | AACACTCACGCAGCTGCT |
| DlTCP19 | Dlo019709 | GCACCGATCACACCAATGT | TGCTACTCGAACTGGAACTGG |
| DlTCP20a | Dlo019007 | GCTGAGCCATCTATAATTGCTG | CACTCCCATAACTTGATGCC |
| DlTCP20b | Dlo027551 | GAAGCCAGCAGAGATCAAAGAT | GGTATGGTTCCACTCCCTGT |
| DlTCP21 | Dlo017839 | GAGAATCCGGATGCCAAT | TTGTGATCCAGTGAAGTCGAG |
| DlTCP23 | Dlo014979 | CCCACCACGAGAACAGTG | GTAAGGAGACGTTAAGGGAGG |
| Fe-SOD | GGTCAGATGGTGAAGCCGTAGAG | GTCTATGCCACCGATACAACAAACCC |
| 基因名称 Gene name | 基因ID Gene ID | 染色 体 Chr | 基因组定位 Gene location | 开放阅 读框 ORF/bp | 氨基酸数 Amino acid number | 分子量 Molecular weight | 理论等 电点 pI | 不稳定 系数 Instability index | 亚细胞定位 Subcellular location |
|---|---|---|---|---|---|---|---|---|---|
| DlTCP1 | Dlo030081 | 14 | 12715538:12717415 | 1 062 | 353 | 39 513.21 | 9.41 | 52.04 | 细胞核Nucleus |
| DlTCP2 | Dlo021979 | 10 | 8735823:8746343 | 1 512 | 503 | 55 328.04 | 8.42 | 54.23 | 细胞核Nucleus |
| DlTCP4a | Dlo009059 | 4 | 10439106:10441967 | 1 251 | 416 | 45 292.48 | 6.35 | 56.10 | 细胞核Nucleus |
| DlTCP4b | Dlo027391 | 13 | 5039037:5042408 | 1 065 | 354 | 38 741.10 | 6.22 | 49.13 | 细胞核Nucleus |
| DlTCP5a | Dlo026162 | 12 | 19976189:19976698 | 510 | 169 | 39 613.57 | 8.62 | 42.30 | 细胞核Nucleus |
| DlTCP5b | Dlo031128 | 15 | 7294709:7295881 | 1 173 | 390 | 43 679.68 | 9.00 | 53.05 | 细胞核Nucleus |
| DlTCP8 | Dlo024507 | 11 | 17156563:17158185 | 1 623 | 540 | 57 211.58 | 6.62 | 61.62 | 细胞核Nucleus |
| DlTCP9 | Dlo012848 | 5 | 32302987:32304165 | 1 179 | 392 | 41 341.56 | 8.97 | 52.88 | 细胞核Nucleus |
| DlTCP11 | Dlo029447 | 14 | 5931490:5932119 | 630 | 209 | 22 462.23 | 8.82 | 74.76 | 细胞核Nucleus |
| DlTCP12 | Dlo018254 | 8 | 18820036:18821397 | 1 362 | 453 | 49 819.29 | 9.10 | 53.43 | 细胞核Nucleus |
| DlTCP13 | Dlo030148 | 14 | 13720270:13723613 | 1 029 | 342 | 37 824.36 | 8.42 | 56.72 | 细胞核Nucleus |
| DlTCP14 | Dlo032223 | 15 | 18165748:18167034 | 1 287 | 428 | 46 492.80 | 7.54 | 63.63 | 细胞核Nucleus |
| DlTCP15 | Dlo018528 | 8 | 21299710:21300948 | 1 239 | 412 | 44 078.51 | 7.35 | 57.80 | 细胞核Nucleus |
| DlTCP17 | Dlo026163 | 12 | 19984109:19984681 | 573 | 190 | 20 595.15 | 6.29 | 37.10 | 叶绿体Chloroplast |
| DlTCP18 | Dlo031253 | 15 | 8846922:8847980 | 1 059 | 352 | 39 608.05 | 7.27 | 49.95 | 细胞核Nucleus |
| DlTCP19 | Dlo019709 | 9 | 5138059:5139069 | 1 011 | 336 | 36 084.09 | 5.9 | 59.25 | 细胞核Nucleus |
| DlTCP20a | Dlo019007 | 8 | 25450145:25450999 | 855 | 284 | 30 303.73 | 9.01 | 44.23 | 细胞核Nucleus |
| DlTCP20b | Dlo027551 | 13 | 6380380:6382053 | 1 152 | 383 | 41 510.79 | 9.06 | 56.77 | 线粒体 Mitochondrion |
| DlTCP21 | Dlo017839 | 8 | 12941326:12942114 | 789 | 262 | 27 391.62 | 9.45 | 39.31 | 细胞核Nucleus |
| DlTCP23 | Dlo014979 | 6 | 25244640:25245548 | 909 | 302 | 32 075.54 | 7.17 | 51.82 | 细胞核Nucleus |
Table 2 Analysis of Physicochemical Properties of DlTCP gene family
| 基因名称 Gene name | 基因ID Gene ID | 染色 体 Chr | 基因组定位 Gene location | 开放阅 读框 ORF/bp | 氨基酸数 Amino acid number | 分子量 Molecular weight | 理论等 电点 pI | 不稳定 系数 Instability index | 亚细胞定位 Subcellular location |
|---|---|---|---|---|---|---|---|---|---|
| DlTCP1 | Dlo030081 | 14 | 12715538:12717415 | 1 062 | 353 | 39 513.21 | 9.41 | 52.04 | 细胞核Nucleus |
| DlTCP2 | Dlo021979 | 10 | 8735823:8746343 | 1 512 | 503 | 55 328.04 | 8.42 | 54.23 | 细胞核Nucleus |
| DlTCP4a | Dlo009059 | 4 | 10439106:10441967 | 1 251 | 416 | 45 292.48 | 6.35 | 56.10 | 细胞核Nucleus |
| DlTCP4b | Dlo027391 | 13 | 5039037:5042408 | 1 065 | 354 | 38 741.10 | 6.22 | 49.13 | 细胞核Nucleus |
| DlTCP5a | Dlo026162 | 12 | 19976189:19976698 | 510 | 169 | 39 613.57 | 8.62 | 42.30 | 细胞核Nucleus |
| DlTCP5b | Dlo031128 | 15 | 7294709:7295881 | 1 173 | 390 | 43 679.68 | 9.00 | 53.05 | 细胞核Nucleus |
| DlTCP8 | Dlo024507 | 11 | 17156563:17158185 | 1 623 | 540 | 57 211.58 | 6.62 | 61.62 | 细胞核Nucleus |
| DlTCP9 | Dlo012848 | 5 | 32302987:32304165 | 1 179 | 392 | 41 341.56 | 8.97 | 52.88 | 细胞核Nucleus |
| DlTCP11 | Dlo029447 | 14 | 5931490:5932119 | 630 | 209 | 22 462.23 | 8.82 | 74.76 | 细胞核Nucleus |
| DlTCP12 | Dlo018254 | 8 | 18820036:18821397 | 1 362 | 453 | 49 819.29 | 9.10 | 53.43 | 细胞核Nucleus |
| DlTCP13 | Dlo030148 | 14 | 13720270:13723613 | 1 029 | 342 | 37 824.36 | 8.42 | 56.72 | 细胞核Nucleus |
| DlTCP14 | Dlo032223 | 15 | 18165748:18167034 | 1 287 | 428 | 46 492.80 | 7.54 | 63.63 | 细胞核Nucleus |
| DlTCP15 | Dlo018528 | 8 | 21299710:21300948 | 1 239 | 412 | 44 078.51 | 7.35 | 57.80 | 细胞核Nucleus |
| DlTCP17 | Dlo026163 | 12 | 19984109:19984681 | 573 | 190 | 20 595.15 | 6.29 | 37.10 | 叶绿体Chloroplast |
| DlTCP18 | Dlo031253 | 15 | 8846922:8847980 | 1 059 | 352 | 39 608.05 | 7.27 | 49.95 | 细胞核Nucleus |
| DlTCP19 | Dlo019709 | 9 | 5138059:5139069 | 1 011 | 336 | 36 084.09 | 5.9 | 59.25 | 细胞核Nucleus |
| DlTCP20a | Dlo019007 | 8 | 25450145:25450999 | 855 | 284 | 30 303.73 | 9.01 | 44.23 | 细胞核Nucleus |
| DlTCP20b | Dlo027551 | 13 | 6380380:6382053 | 1 152 | 383 | 41 510.79 | 9.06 | 56.77 | 线粒体 Mitochondrion |
| DlTCP21 | Dlo017839 | 8 | 12941326:12942114 | 789 | 262 | 27 391.62 | 9.45 | 39.31 | 细胞核Nucleus |
| DlTCP23 | Dlo014979 | 6 | 25244640:25245548 | 909 | 302 | 32 075.54 | 7.17 | 51.82 | 细胞核Nucleus |
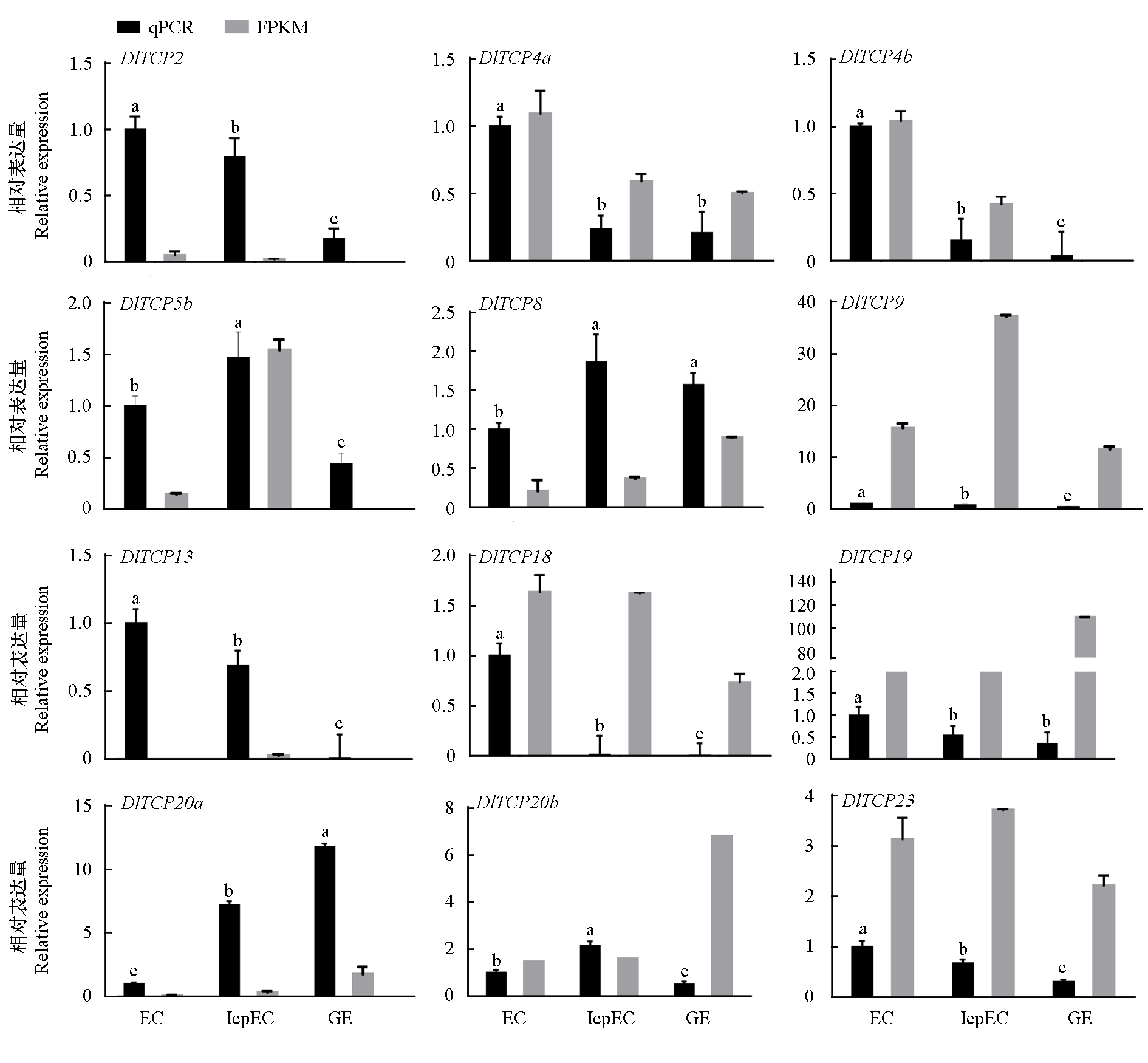
Fig. 8 Analysis of the qRT-PCR of ">DlTCP genes family in the early somatic embryogenesis of longan EC:Embryogenic callus;IcpEC:Incomplete compact pro-embrogenic culture;GE:Globular embryos.Different lowercase letters indicate significant differences between data.
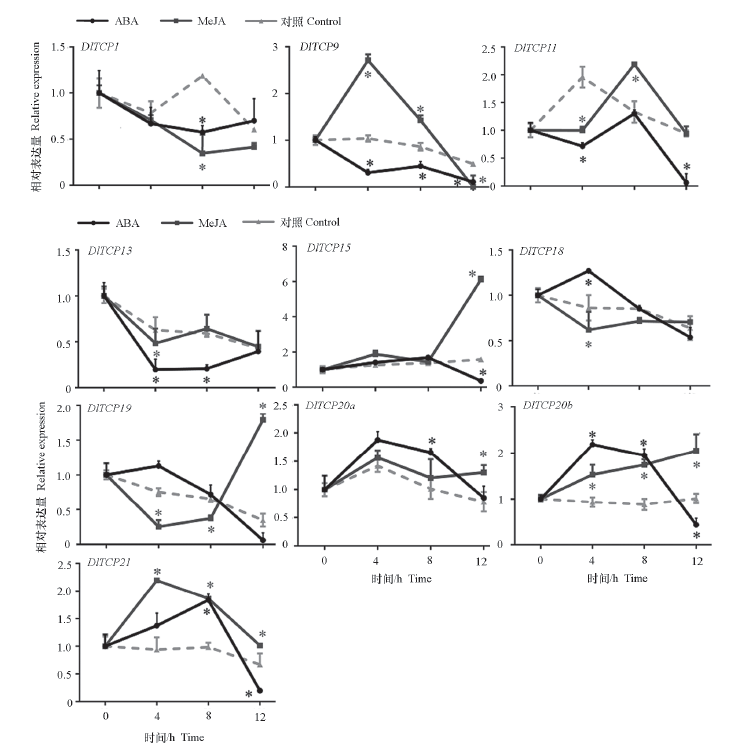
Fig. 9 Expression levels of 10 selected DlTCP in longan EC after ABA and MeJA treatments ABA and MeJA were significantly analyzed with control respectively,* indicates that there is significant difference between the two groups of data.
| [1] | Chai W B, Jiang P F, Guo Y, Huang H Y, Li X Y. 2017. Identification and expression profiling analysis of TCP family genes involved in growth and development in maize. Physiology & Molecular Biology of Plants, 23:779-791. |
| [2] |
Chen C J, Chen H, Zhang Y, Thomas H R, Frank M H, He Y H, Xia R. 2020. TBtools:an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 13 (8):1194-1202.
doi: 10.1016/j.molp.2020.06.009 URL |
| [3] |
Cubas P, Lauter N, Doebley J, Coen E. 1999. The TCP domain:a motif found in proteins regulating plant growth and development. Plant J, 18 (2):215-222.
pmid: 10363373 |
| [4] |
Danisman S, van Dijk A D J, Bimbo A, van der Wal F, Hennig L, de Folter S, Angenent G C, Immink R G H. 2013. Analysis of functional redundancies within the Arabidopsis TCP transcription factor family. Journal of Experimental Botany, 64 (18):5673-5685.
doi: 10.1093/jxb/ert337 URL |
| [5] |
Doebley J, Stec A, Hubbard L. 1997. The evolution of apical dominance in maize. Nature, 386 (6624):485-488.
doi: 10.1038/386485a0 URL |
| [6] |
Guo Z, Fujioka S, Blancaflor E, Miao S, Gou X, Li J. 2010. TCP 1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell, 22 (4):1161-1173.
doi: 10.1105/tpc.109.069203 URL |
| [7] | Kieffer M, Master V, Waites R, Davies B. 2011. TCP14 and TCP 15 affect internode length and leaf shape in Arabidopsis. Plant Journal for Cell & Molecular Biology, 68 (1):147-158. |
| [8] | Kosugi S, Ohashi Y. 1997. PCF1 and PCF 2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. The Plant Cell, 9 (9):1607-1619. |
| [9] |
Koyama T, Mitsuda N, Seki M, Shinozaki K, Ohmetakagi M. 2010. TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164,as well as the auxin response,during differentiation of leaves in Arabidopsis. Plant Cell, 22 (11):3574.
doi: 10.1105/tpc.110.075598 URL |
| [10] | Lai Zhong-xiong. 2003. Study of longan biotechnology. Fuzhou: Fujian Science and Technology Press. (in Chinese) |
| 赖钟雄. 2003. 龙眼生物技术研究. 福州: 福建科学技术出版社. | |
| [11] |
Leng X, Wei H, Xu X, Ghuge S A, Jia D, Liu G, Wang Y, Yuan Y. 2019. Genome-wide identification and transcript analysis of TCP transcription factors in grapevine. BMC Genomics, 20 (1):786.
doi: 10.1186/s12864-019-6159-2 URL |
| [12] | Li S. 2015. The Arabidopsis thaliana TCP transcription factors:a broadening horizon beyond development. Plant Signaling & Behavior, 10 (7):e1044192. |
| [13] |
Li S, Zachgo S. 2013. TCP 3 interacts with R2R3-MYB proteins,promotes flavonoid biosynthesis and negatively regulates the auxin response in Arabidopsis thaliana. Plant Journal, 76 (6):901-913.
doi: 10.1111/tpj.2013.76.issue-6 URL |
| [14] |
Lin Y F, Chen Y Y, Yu-Yun H, Shen C Y, Jui-Ling H, Chuan-Ming Y, Nobutaka M, Masaru O T, Liu Z J, Wen-Chieh T. 2016. Genome-wide identification and characterization of TCP genes involved in ovule development of Phalaenopsis equestris. Journal of Experimental Botany, 67 (17):5051-5066.
doi: 10.1093/jxb/erw273 pmid: 27543606 |
| [15] |
Lin Y L, Lai Z X. 2010. Reference gene selection for qPCR analysis during somatic embryogenesis in longan tree. Plant Science, 178 (4):359-365.
doi: 10.1016/j.plantsci.2010.02.005 URL |
| [16] | Lin Y L, Min J M, Lai R L, Wu Z Y, Chen Y K, Yu L L, Cheng C Z, Jin Y C, Tian Q L, Liu Q F. 2017. Genome-wide sequencing of longan (Dimocarpus longan Lour.)provides insights into molecular basis of its polyphenol-rich characteristics. Gigaence, 6 (5):1-14. |
| [17] |
Liu H, Gao Y, Wu M, Shi Y, Wang H, Wu L, Xiang Y. 2020. TCP10,a TCP transcription factor in moso bamboo(Phyllostachys edulis),confers drought tolerance to transgenic plants. Environmental and Experimental Botany, 172 (9):104002.
doi: 10.1016/j.envexpbot.2020.104002 URL |
| [18] |
Liu Y, Li D, Yan J, Wang K, Luo H, Zhang W. 2019. MiR319 mediated salt tolerance by ethylene biosynthesis,signaling and salt stress response in switchgrass. Plant Biotechnology Journal, 17 (12):2370-2383.
doi: 10.1111/pbi.v17.12 URL |
| [19] |
Lopez J A, Sun Y, Blair P B, Mukhtar M S. 2015. TCP three-way handshake:linking developmental processes with plant immunity. Trends in Plant Science, 20 (4):238-245.
doi: 10.1016/j.tplants.2015.01.005 URL |
| [20] |
Luo D, Carpenter R, Vincent C, Copsey L, Coen E. 1996. Origin of floral asymmetry in Antirrhinum. Nature, 383 (6603):794-799.
doi: 10.1038/383794a0 URL |
| [21] | Ma J, Wang Q, Sun R, Xie F, Jones D C, Zhang B. 2014. Genome-wide identification and expression analysis of TCP transcription factors in Gossypium raimondii. Rep, 4:6645. |
| [22] | Martín-Trillo M, Cubas P. 2010. TCP genes:a family snapshot ten years later. Trends in Plant Science, 15 (1):39. |
| [23] | Min L, Hu Q, Li Y, Xu J, Ma Y, Zhu L, Yang X, Zhang X. 2015. Leafy cotyledon1-casein kinaseⅠ-TCP15-phytochome interacting factor 4 network regulates somatic embryogenesis by regulating auxin homeostasis. Plant Physiology, 169 (4):1480-2015. |
| [24] | Nag A, King S, Jack T. 2009. miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proceedings of the National Academy of Sciences, 106 (52):22534-22539. |
| [25] |
Olivier N, Patrick D, Elodie C, Dominique T, Christine H. 2007. TCP transcription factors predate the emergence of land plants. Journal of Molecular Evolution, 65 (1):23-33.
pmid: 17568984 |
| [26] |
Om P G, Anil D, Archana S, Sweta K, Pradeep K J T, Vinutha&Shelly P. 2019. Conserved miRNAs modulate the expression of potential transcription factors of isoflavonoid biosynthetic pathway in soybean seeds. Molecular Biology Reports, 46 (4):3713-3730.
doi: 10.1007/s11033-019-04814-7 URL |
| [27] |
Parapunova V, Busscher M, Busscher-Lange J, Lammers M, Karlova R, Bovy A G, Angenent G C, de Maagd R A. 2014. Identification,cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biology, 14:157.
doi: 10.1186/1471-2229-14-157 pmid: 24903607 |
| [28] |
Resentini F, Felipo-Benavent A, Colombo L, Blázquez M A, Alabadi D, Masiero S. 2015. TCP14 and TCP15 mediate the promotion of seed germination by gibberellins in Arabidopsis thaliana. Molecular Plant, 8 (3):482-485.
doi: 10.1016/j.molp.2014.11.018 URL |
| [29] | Sarvepalli K, Nath U. 2011. Hyper-activation of the TCP 4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. Plant Journal for Cell & Molecular Biology, 67 (4):595-607. |
| [30] |
Schommer C, Debernardi J M, Bresso E G, Rodriguez R E, Palatnik J F. 2014. Repression of cell proliferation by miR319-regulated TCP4. Molecular Plant, 7 (10):1533-1544.
doi: 10.1093/mp/ssu084 pmid: 25053833 |
| [31] |
Schommer C, Palatnik J F, Aggarwal P, Chételat A, Cubas P, Farmer E E, Nath U, Weigel D, Carrington J C. 2008. Control of jasmonate biosynthesis and senescence by miR319 Targets. PLoS Biology, 6 (9):e230.
doi: 10.1371/journal.pbio.0060230 URL |
| [32] |
Shi P, Guy K M, Wu W, Fang B, Yang J, Zhang M, Hu Z. 2016. Genome-wide identification and expression analysis of the ClTCP transcription factors in Citrullus lanatus. BMC Plant Biology, 16 (1):85.
doi: 10.1186/s12870-016-0765-9 URL |
| [33] | Su Li-yao, Chen Xu, Huang Shu-qi, Jiang Meng-qi, Li Xue, Zhang Zi-hao, Lai Zhong-xiong, Lin Yu-ling. 2020. The expression profiles of eTM,microRNA319 and their targets in early longan somatic embryogenesis. Chinese Journal of Applied & Environmental Biology, 26 (3):566-573. (in Chinese) |
| 苏立遥, 陈旭, 黄倏祺, 蒋梦琦, 厉雪, 张梓浩, 赖钟雄, 林玉玲. 2020. eTM、microRNA319及其调控靶标在龙眼体胚发生早期的表达模式. 应用与环境生物学报, 26 (3):566-573. | |
| [34] |
Sun X, Wang C, Nan X, Xiong L, Yang Y. 2017. Activation of secondary cell wall biosynthesis by miR319-targeted TCP 4 transcription factor. Plant Biotechnology Journal, 15 (10):1284.
doi: 10.1111/pbi.2017.15.issue-10 URL |
| [35] |
Takeda T, Amano K, Ohto M A, Nakamura K, Sato S, Kato T, Tabata S, Ueguchi C. 2006. RNA interference of the Arabidopsis putative transcription factor TCP 16 gene results in abortion of early pollen development. Plant Molecular Biology, 61 (1-2):165-177.
pmid: 16786299 |
| [36] |
Tran C D, Chu H D, Nguyen K H, Watanabe Y, La H V, Tran K D, Tran L P. 2018. Genome-wide identification of the TCP transcription factor family in Chickpea(Cicer arietinum L.)and their transcriptional responses to dehydration and exogenous abscisic acid treatments. Journal of Plant Growth Regulation, 37 (4):1286-1299.
doi: 10.1007/s00344-018-9859-y URL |
| [37] | Wen Beibei, Luo Yong, Liu Dongmin, Zhang Xiangna, Li Juan, Wang Yingzi, Wang Kunbo, Huang Jianan. 2019. Identification and expression profiling analysis of TCP family genes involved in growth and development in Camellia sinensis. Acta Horticulturae Sinica, 46 (12):2369-2382. (in Chinese) |
| 温贝贝, 罗勇, 刘冬敏, 张向娜, 李娟, 王英姿, 王坤波, 黄建安. 2019. 茶树TCP转录因子的鉴定与表达分析. 园艺学报, 46 (12):2369-2382. | |
| [38] |
Wu Z, Wang W, Zhuang J. 2017. TCP family genes control leaf development and its responses to hormonal stimuli in tea plant [Camellia sinensis(L.)O. Kuntze]. Plant Growth Regulation, 83 (1):43-53.
doi: 10.1007/s10725-017-0282-3 URL |
| [39] |
Yao X, Hong M, Jian W. 2010. Genome-wide comparative analysis and expression pattern of TCP gene families in Arabidopsis thaliana and Oryza sativa. Journal of Integrative Plant Biology, 49 (6):885-897.
doi: 10.1111/jipb.2007.49.issue-6 URL |
| [40] |
Zhang X, Bao Y, Shan D, Wang Z, Song X, Wang Z, Wang J, He L, Wu L, Zhang Z, Niu D, Jin H, Zhao H. 2018. Magnaporthe oryzae induces the expression of a microRNA to suppress the immune response in rice. Plant Physiology, 177 (1):352-368.
doi: 10.1104/pp.17.01665 pmid: 29549093 |
| [1] | YUAN Xin, XU Yunhe, ZHANG Yupei, SHAN Nan, CHEN Chuying, WAN Chunpeng, KAI Wenbin, ZHAI Xiawan, CHEN Jinyin, GAN Zengyu. Studies on AcAREB1 Regulating the Expression of AcGH3.1 During Postharvest Ripening of Kiwifruit [J]. Acta Horticulturae Sinica, 2023, 50(1): 53-64. |
| [2] | SUN Simiao, WANG Kun, GAO Yuan, WANG Dajiang, and LI Lianwen. A New Ornamental Crabapple Cultivar‘Zichen’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 267-268. |
| [3] | ZHANG Qiuyue, LIU Changlai, YU Xiaojing, YANG Jiading, FENG Chaonian. Screening of Reference Genes for Differentially Expressed Genes in Pyrus betulaefolia Plant Under Salt Stress by qRT-PCR [J]. Acta Horticulturae Sinica, 2022, 49(7): 1557-1570. |
| [4] | WANG Hong, YANG Wangli, LIN Jing, YANG Qingsong, LI Xiaogang, SHENG Baolong, CHANG Youhong. Comparative Metabolic and Transcriptomic Analysis of Ripening Fruit in Pear Cultivars of‘Sucui 1’‘Cuiguan’and‘Huasu’ [J]. Acta Horticulturae Sinica, 2022, 49(3): 493-508. |
| [5] | LIU Mengyu, JIANG Mengqi, CHEN Yan, ZHANG Shuting, XUE Xiaodong, XIAO Xuechen, LAI Zhongxiong, LIN Yuling. Genome-wide Identification and Expression Analysis of GDSL Esterase/Lipase Genes in Dimocarpus longan [J]. Acta Horticulturae Sinica, 2022, 49(3): 597-612. |
| [6] | SHEN Xu, CHEN Xiaohui, ZHANG Jing, CHEN Rongzhu, XU Xiaoping, LI Xiaofei, JIANG Mengqi, LIU Pudong, NI Shanshan, LIN Yuling, LAI Zhongxiong. Evolutionary Dynamics Investigation and the Expression Analysis of Chromatin Remodeling Factor Snf2 Gene Family During Early Somatic Embryogenesis in Dimocarpus longan [J]. Acta Horticulturae Sinica, 2022, 49(1): 41-61. |
| [7] | XIE Siyi, ZHOU Chengzhe, ZHU Chen, ZHAN Dongmei, CHEN Lan, WU Zuchun, LAI Zhongxiong, GUO Yuqiong. Genome-wide Identification and Expression Analysis of CsTIFY Transcription Factor Family Under Abiotic Stress and Hormone Treatments in Camellia sinensis [J]. Acta Horticulturae Sinica, 2022, 49(1): 100-116. |
| [8] | A New Chaenomeles Crabapple Cultivar‘Caiyu’. A New Chaenomeles Crabapple Cultivar‘Caiyu’ [J]. Acta Horticulturae Sinica, 2021, 48(S2): 2975-2976. |
| [9] | ZHOU Jinchuan, GUAN Qide, GUAN Xueying, ZHAO Yuhan, ZHANG Haijuan, and WANG Xiangbo. A New Chaenomeles Crabapple Cultivar‘Zui Xishi’ [J]. Acta Horticulturae Sinica, 2021, 48(S2): 2977-2978. |
| [10] | ZHANG Zhifang, YUAN lei, CHENG Nini, LIU Zongzhao, and CHEN Zhiqun, . A New Ornamental Crabapple Cultivar‘Fengguanhong’ [J]. Acta Horticulturae Sinica, 2021, 48(S2): 2979-2980. |
| [11] | CHEN Li, ZHOU Jinchuan, HAN Qingdian, WANG Xiangbo, GUAN Qide, and CHEN Zhiqun, . A New Crabapple Cultivar‘Menghuijuan’ [J]. Acta Horticulturae Sinica, 2021, 48(S2): 2981-2982. |
| [12] | ZHANG Haijuan, GUAN Qide, ZHOU Jinchuan, and GAO Yueheng. A New Chaenomeles Crabapple Cultivar‘Huatiannü’ [J]. Acta Horticulturae Sinica, 2021, 48(S2): 2983-2984. |
| [13] | ZHANG Chunyu, XU Xiaoqiong, XU Xiaoping, ZHAO Pengcheng, SHEN Xu, Munir Nigarish, ZHANG Zihao, LIN Yuling, Chen Zhenguang, LAI Zhongxiong. Genome-wide Identification of the SKP1-like Family and Analysis of Their Expression During Early Somatic Embryogenesis in Longan [J]. Acta Horticulturae Sinica, 2021, 48(9): 1665-1679. |
| [14] | ZHANG Zhiqiang, LU Shixiong, MA Zonghuan, LI Yanbiao, GAO Caixia, CHEN Baihong, MAO Juan. Bioinformatic Identification and Expression Analysis of Candidate Genes of LIM Protein Family in Strawberry Under Abiotic Stresses [J]. Acta Horticulturae Sinica, 2021, 48(8): 1485-1503. |
| [15] | ZHU Junfei, LI Xin, DONG Kangting, TANG Zhifei, BIAN Xiuju, WANG Lihong, LI Huibin, SUN Xinbo. Effect of Creeping Bentgrass Small Heat Shock Protein AsHSP26.8a on Transgenic Photosynthesis in Arabidopsis thaliana [J]. Acta Horticulturae Sinica, 2021, 48(8): 1619-1625. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd