
Acta Horticulturae Sinica ›› 2021, Vol. 48 ›› Issue (11): 2211-2226.doi: 10.16420/j.issn.0513-353x.2020-0913
• Original article • Previous Articles Next Articles
WANG Peng1, TIAN Zhejuan1, KANG Chen1, LI Yadong1, WANG Hongle2, YANG Chaosha1, MANG Guangwei2, KANG Liang2, FAN Qingjie2, WU Zhiming1,*( )
)
Received:2021-05-24
Revised:2021-08-06
Published:2021-12-02
Contact:
WU Zhiming
E-mail:zhiming71@126.com
CLC Number:
WANG Peng, TIAN Zhejuan, KANG Chen, LI Yadong, WANG Hongle, YANG Chaosha, MANG Guangwei, KANG Liang, FAN Qingjie, WU Zhiming. Establishment and Application of a Tomato KASP Genotyping System Based on Five Disease Resistance Genes[J]. Acta Horticulturae Sinica, 2021, 48(11): 2211-2226.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2020-0913
| 抗病基因 Gene | 引物名称 Primer name | 引物序列(5′-3′) Primer sequence | 抗病标记/bp Resisting marker | 感病标记/bp Unresisting marker | 参考文献 Reference |
|---|---|---|---|---|---|
| Ty-1 | NL3-F | TGTATGCGTCGTGTTAGAAGGTC | 984 | 1 434 | 葛乃蓬 等, |
| NL3-R | AATGGTGACAGAATCAAGATCCAC | ||||
| NL2-F | AAACGCTCTTCTTCCCCTCG | ||||
| NL2-R | AGTTAGATTCAGGCTCCTCGCA | ||||
| Mi-1 | PMiF3 | GGTATGAGCATGCTTAATCAGAGCTCTC | 550 | 350 | Arens et al., |
| PMiR3 | CCTACAAGAAATTATTGTGCGTGTGAATG | ||||
| Tm-22 | Tm-2a-A1 | GCCTCATTCAACTTCCTTCTGG | 472 | 朱明涛 等, | |
| Tm-2a-A2 | GCCAGTATATAACGGTCTAAAC | ||||
| Frl | C2-25-F | ATGGGCGCTGCATGTTTCGTG | 750 | 1 000 | Staniaszek et al., |
| C2-25-R | ACACCTTTGTTGAAAGCCATCCC | ||||
| Sw-5 | Sw-5-2F | AATTAGGTTCTTGAAGCCCATCT | 574 | 510 | Dianese et al., |
| Sw-5-2R | TTCCGCATCAGCCAATAGTGT |
Table 1 PCR primers used in this study
| 抗病基因 Gene | 引物名称 Primer name | 引物序列(5′-3′) Primer sequence | 抗病标记/bp Resisting marker | 感病标记/bp Unresisting marker | 参考文献 Reference |
|---|---|---|---|---|---|
| Ty-1 | NL3-F | TGTATGCGTCGTGTTAGAAGGTC | 984 | 1 434 | 葛乃蓬 等, |
| NL3-R | AATGGTGACAGAATCAAGATCCAC | ||||
| NL2-F | AAACGCTCTTCTTCCCCTCG | ||||
| NL2-R | AGTTAGATTCAGGCTCCTCGCA | ||||
| Mi-1 | PMiF3 | GGTATGAGCATGCTTAATCAGAGCTCTC | 550 | 350 | Arens et al., |
| PMiR3 | CCTACAAGAAATTATTGTGCGTGTGAATG | ||||
| Tm-22 | Tm-2a-A1 | GCCTCATTCAACTTCCTTCTGG | 472 | 朱明涛 等, | |
| Tm-2a-A2 | GCCAGTATATAACGGTCTAAAC | ||||
| Frl | C2-25-F | ATGGGCGCTGCATGTTTCGTG | 750 | 1 000 | Staniaszek et al., |
| C2-25-R | ACACCTTTGTTGAAAGCCATCCC | ||||
| Sw-5 | Sw-5-2F | AATTAGGTTCTTGAAGCCCATCT | 574 | 510 | Dianese et al., |
| Sw-5-2R | TTCCGCATCAGCCAATAGTGT |
| 抗病基因 Gene | 引物名称 Primer name | 引物序列(5′-3′) Primer sequence | 等位基因 Allele | 参考文献 Reference |
|---|---|---|---|---|
| Ty-1 | Ty-1-Allele-F1 | GAAGGTCGGAGTCAACGGATTGAATATGGTAAAGGTGCATCCAGA | A︰A | 胡霞 等, |
| Ty-1-Allele-F2 | GAAGGTGACCAAGTTCATGCTGAATATGGTAAAGGTGCATCCAGT | T︰T | ||
| Ty-1-R | CCGTTGATTGAAGAAATTGATG | |||
| Mi-1 | Mi-1-Allele-R1 | GAAGGTGACCAAGTTCATGCTGATGTTTACAAGTATTTGTGGCG | G︰G | Zübeyir et al., |
| Mi-1-Allele-R2 | GAAGGTCGGAGTCAACGGATTGATGTTTACAAGTATTTGTGGCA | A︰A | ||
| Mi-1-F | TTCAGACGAAGTTAGACATAAAGGA | |||
| Tm-22 | Tm-2-Allele-F1 | GAAGGTCGGAGTCAACGGATTCAATATGCTACAAGCCAAATTACTC | C︰C | 孔会利 等, |
| Tm-2-Allele-F2 | GAAGGTGACCAAGTTCATGCTCAATATGCTACAAGCCAAATTACTT | T︰T | ||
| Tm-2-R | GAAACAACTCCATAATGCAAATC | |||
| Frl | Frl-Allele-F1 | GAAGGTGACCAAGTTCATGCTTAAAGGACCATTTTTCGTAGACCATT | T︰T | 国家进 等, |
| Frl-Allele-F2 | GAAGGTCGGAGTCAACGGATTTAAAGGACCATTTTTCGTAGACCATA | A︰A | ||
| Frl-R | ATTTCTTACCATATGTGAAGAGAA | |||
| Sw-5 | FP1SNP724 | GAAGGTCGGAGTCAACGGATTCATGGACACAACTGGAGTTATTA | A︰A | Devran & Kahveci, |
| FP2SNP724 | GAAGGTGACCAAGTTCATGCTCATGGACACAACTGGAGTTATTG | G︰G | ||
| RPSNP724 | CAGTCATTCTTCAAATTTTACTTGT |
Table 2 KASP primers used in this study
| 抗病基因 Gene | 引物名称 Primer name | 引物序列(5′-3′) Primer sequence | 等位基因 Allele | 参考文献 Reference |
|---|---|---|---|---|
| Ty-1 | Ty-1-Allele-F1 | GAAGGTCGGAGTCAACGGATTGAATATGGTAAAGGTGCATCCAGA | A︰A | 胡霞 等, |
| Ty-1-Allele-F2 | GAAGGTGACCAAGTTCATGCTGAATATGGTAAAGGTGCATCCAGT | T︰T | ||
| Ty-1-R | CCGTTGATTGAAGAAATTGATG | |||
| Mi-1 | Mi-1-Allele-R1 | GAAGGTGACCAAGTTCATGCTGATGTTTACAAGTATTTGTGGCG | G︰G | Zübeyir et al., |
| Mi-1-Allele-R2 | GAAGGTCGGAGTCAACGGATTGATGTTTACAAGTATTTGTGGCA | A︰A | ||
| Mi-1-F | TTCAGACGAAGTTAGACATAAAGGA | |||
| Tm-22 | Tm-2-Allele-F1 | GAAGGTCGGAGTCAACGGATTCAATATGCTACAAGCCAAATTACTC | C︰C | 孔会利 等, |
| Tm-2-Allele-F2 | GAAGGTGACCAAGTTCATGCTCAATATGCTACAAGCCAAATTACTT | T︰T | ||
| Tm-2-R | GAAACAACTCCATAATGCAAATC | |||
| Frl | Frl-Allele-F1 | GAAGGTGACCAAGTTCATGCTTAAAGGACCATTTTTCGTAGACCATT | T︰T | 国家进 等, |
| Frl-Allele-F2 | GAAGGTCGGAGTCAACGGATTTAAAGGACCATTTTTCGTAGACCATA | A︰A | ||
| Frl-R | ATTTCTTACCATATGTGAAGAGAA | |||
| Sw-5 | FP1SNP724 | GAAGGTCGGAGTCAACGGATTCATGGACACAACTGGAGTTATTA | A︰A | Devran & Kahveci, |
| FP2SNP724 | GAAGGTGACCAAGTTCATGCTCATGGACACAACTGGAGTTATTG | G︰G | ||
| RPSNP724 | CAGTCATTCTTCAAATTTTACTTGT |
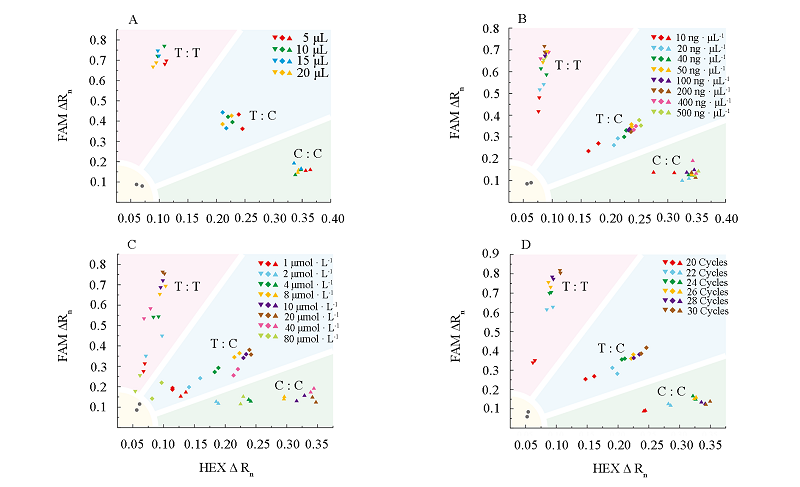
Fig. 2 KASP genotyping with different reaction volumes(A),different concentrations of template DNA(B),different primer concentrations(C),different number of reaction cycles(D)for Tm-22 gene
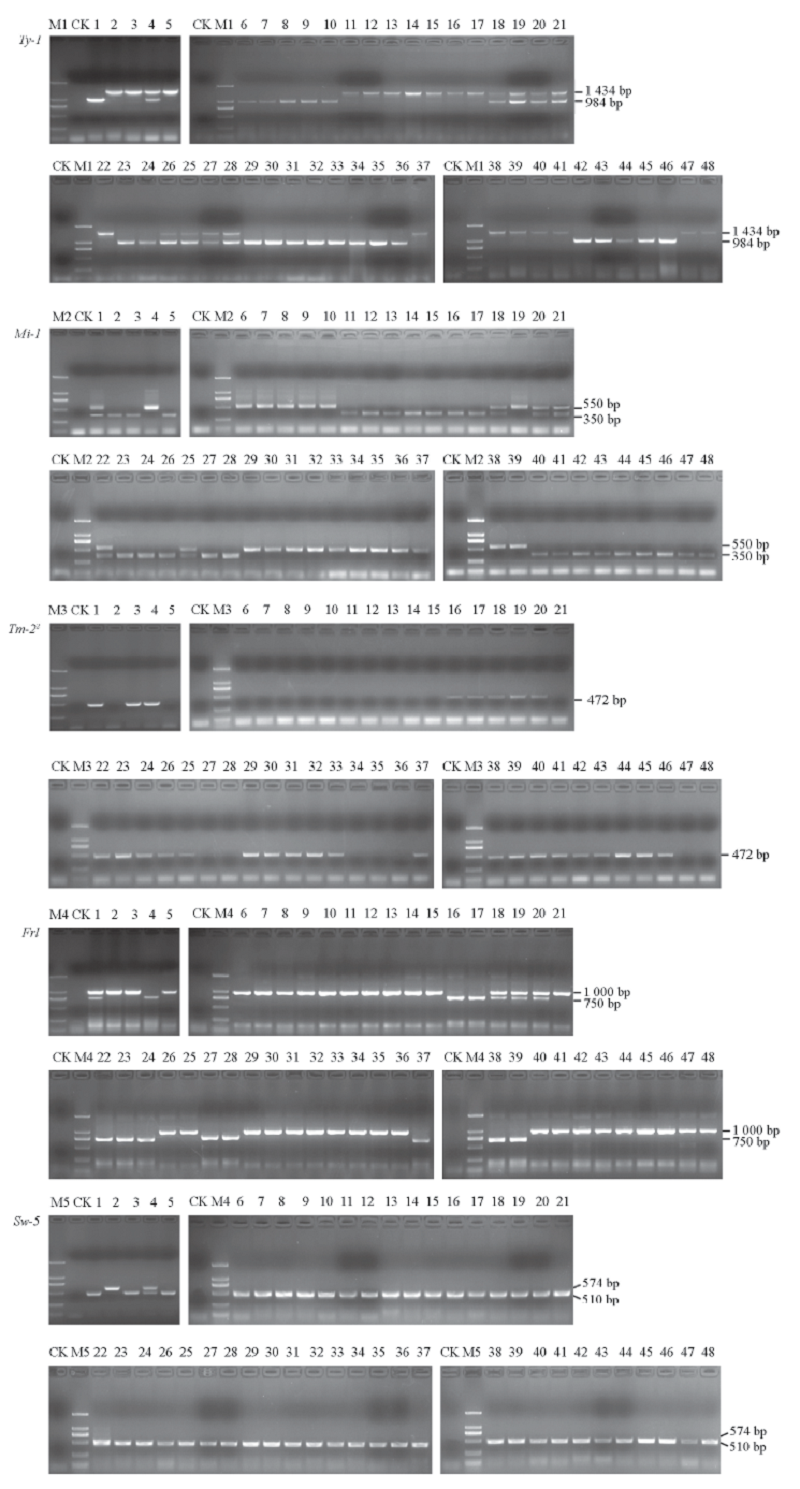
Fig. 3 PCR detection of resistance genes Ty-1,Mi-1,Tm-22,Frl and Sw-5 CK:Negative control;M1 ~ M5:Marker 100 bp,250 bp,500 bp,750 bp,1 000 bp,2 000 bp;1-48:Materials of tomato.
| 编号 No. | Ty-1 | Mi-1 | Tm-22 | Frl | Sw-5 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCR | KASP | 基因型 Genotype | PCR | KASP | 基因型 Genotype | PCR | KASP | 基因型 Genotype | PCR | KASP | 基因型 Genotype | PCR | KASP | 基因型 Genotype | |
| 1 | ++ | A︰A | Ty-1/Ty-1 | +- | G︰A | Mi-1/mi-1 | + | T︰C | Tm-22/tm-22 | + - | A︰T | Frl/frl | -- | A︰A | sw-5/sw-5 |
| 2 | - - | T︰T | ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | + + | G︰G | Sw-5/Sw-5 |
| 3 | - - | T︰T | ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | + | T︰T | Tm-22/Tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 4 | + - | A︰T | Ty-1/ty-1 | ++ | G︰G | Mi-1/Mi-1 | + | T︰C | Tm-22/tm-22 | + + | A︰A | Frl/Frl | +- | G︰A | Sw-5/sw-5 |
| 5 | - - | T︰T | ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 6 | ++ | A︰A | Ty-1/Ty-1 | + + | G︰G | Mi-1/Mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 7 | ++ | A︰A | Ty-1/Ty-1 | + + | G︰G | Mi-1/Mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 8 | ++ | A︰A | Ty-1/Ty-1 | + + | G︰G | Mi-1/Mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 9 | + + | A︰A | Ty-1/Ty-1 | + + | G︰G | Mi-1/Mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 10 | + + | A︰A | Ty-1/Ty-1 | + + | G︰G | Mi-1/Mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 11 | - - | T︰T | ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 12 | - - | T︰T | ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 13 | - - | T︰T | ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 14 | - - | T︰T | ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 15 | - - | T︰T | ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 16 | - - | T︰T | ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | + | T︰T | Tm-22/Tm-22 | + + | A︰A | Frl/Frl | -- | A︰A | sw-5/sw-5 |
| 17 | - - | T︰T | ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | + | T︰T | Tm-22/Tm-22 | + + | A︰A | Frl/Frl | -- | A︰A | sw-5/sw-5 |
| 18 | + - | A︰T | Ty-1/ty-1 | +- | G︰A | Mi-1/mi-1 | + | T︰C | Tm-22/tm-22 | + - | A︰T | Frl/frl | -- | A︰A | sw-5/sw-5 |
| 19 | + - | A︰T | Ty-1/ty-1 | + + | G︰G | Mi-1/Mi-1 | + | T︰C | Tm-22/tm-22 | + - | A︰T | Frl/frl | -- | A︰A | sw-5/sw-5 |
| 20 | + - | A︰T | Ty-1/ty-1 | +- | G︰A | Mi-1/mi-1 | + | T︰C | Tm-22/tm-22 | + - | A︰T | Frl/frl | -- | A︰A | sw-5/sw-5 |
| 21 | + - | A︰T | Ty-1/ty-1 | +- | G︰A | Mi-1/mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 22 | - - | T︰T | ty-1/ty-1 | +- | G︰A | Mi-1/mi-1 | + | T︰T | Tm-22/Tm-22 | + + | A︰A | Frl/Frl | -- | A︰A | sw-5/sw-5 |
| 23 | + + | A︰A | Ty-1/Ty-1 | -- | A︰A | mi-1/mi-1 | + | T︰T | Tm-22/Tm-22 | + + | A︰A | Frl/Frl | -- | A︰A | sw-5/sw-5 |
| 24 | + + | A︰A | Ty-1/Ty-1 | -- | A︰A | mi-1/mi-1 | + | T︰T | Tm-22/Tm-22 | + + | A︰A | Frl/Frl | -- | A︰A | sw-5/sw-5 |
| 25 | + - | A︰T | Ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | + | T︰C | Tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 26 | + - | A︰T | Ty-1/ty-1 | +- | G︰A | Mi-1/mi-1 | + | T︰C | Tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 27 | + - | A︰T | Ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | - - | C︰C | tm-22/tm-22 | + + | A︰A | Frl/Frl | -- | A︰A | sw-5/sw-5 |
| 28 | + - | A︰T | Ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | - - | C︰C | tm-22/tm-22 | + + | A︰A | Frl/Frl | -- | A︰A | sw-5/sw-5 |
| 29 | + + | A︰A | Ty-1/Ty-1 | + + | G︰G | Mi-1/Mi-1 | + | T︰T | Tm-22/Tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 30 | + + | A︰A | Ty-1/Ty-1 | + + | G︰G | Mi-1/Mi-1 | + | T︰T | Tm-22/Tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 31 | + + | A︰A | Ty-1/Ty-1 | + + | G︰G | Mi-1/Mi-1 | + | T︰T | Tm-22/Tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 32 | + + | A︰A | Ty-1/Ty-1 | + + | G︰G | Mi-1/Mi-1 | + | T︰T | Tm-22/Tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 33 | + + | A︰A | Ty-1/Ty-1 | + + | G︰G | Mi-1/Mi-1 | + | T︰T | Tm-22/Tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 34 | + + | A︰A | Ty-1/Ty-1 | + + | G︰G | Mi-1/Mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 35 | + + | A︰A | Ty-1/Ty-1 | + + | G︰G | Mi-1/Mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 36 | + + | A︰A | Ty-1/Ty-1 | + + | G︰G | Mi-1/Mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 37 | - - | T︰T | ty-1/ty-1 | + + | G︰G | Mi-1/Mi-1 | + | T︰T | Tm-22/Tm-22 | + + | A︰A | Frl/Frl | -- | A︰A | sw-5/sw-5 |
| 38 | - - | T︰T | ty-1/ty-1 | + + | G︰G | Mi-1/Mi-1 | + | T︰T | Tm-22/Tm-22 | + + | A︰A | Frl/Frl | -- | A︰A | sw-5/sw-5 |
| 39 | - - | T︰T | ty-1/ty-1 | + + | G︰G | Mi-1/Mi-1 | + | T︰T | Tm-22/Tm-22 | + + | A︰A | Frl/Frl | -- | A︰A | sw-5/sw-5 |
| 40 | - - | T︰T | ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | + | T︰T | Tm-22/Tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 41 | - - | T︰T | ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | + | T︰T | Tm-22/Tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 42 | + + | A︰A | Ty-1/Ty-1 | -- | A︰A | mi-1/mi-1 | + | T︰T | Tm-22/Tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 43 | + + | A︰A | Ty-1/Ty-1 | -- | A︰A | mi-1/mi-1 | + | T︰T | Tm-22/Tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 44 | + + | A︰A | Ty-1/Ty-1 | -- | A︰A | mi-1/mi-1 | + | T︰T | Tm-22/Tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 45 | + + | A︰A | Ty-1/Ty-1 | -- | A︰A | mi-1/mi-1 | + | T︰T | Tm-22/Tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 46 | + + | A︰A | Ty-1/Ty-1 | -- | A︰A | mi-1/mi-1 | + | T︰T | Tm-22/Tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 47 | - - | T︰T | ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 48 | - - | T︰T | ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
Table 3 Identification results of tomato disease resistance
| 编号 No. | Ty-1 | Mi-1 | Tm-22 | Frl | Sw-5 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCR | KASP | 基因型 Genotype | PCR | KASP | 基因型 Genotype | PCR | KASP | 基因型 Genotype | PCR | KASP | 基因型 Genotype | PCR | KASP | 基因型 Genotype | |
| 1 | ++ | A︰A | Ty-1/Ty-1 | +- | G︰A | Mi-1/mi-1 | + | T︰C | Tm-22/tm-22 | + - | A︰T | Frl/frl | -- | A︰A | sw-5/sw-5 |
| 2 | - - | T︰T | ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | + + | G︰G | Sw-5/Sw-5 |
| 3 | - - | T︰T | ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | + | T︰T | Tm-22/Tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 4 | + - | A︰T | Ty-1/ty-1 | ++ | G︰G | Mi-1/Mi-1 | + | T︰C | Tm-22/tm-22 | + + | A︰A | Frl/Frl | +- | G︰A | Sw-5/sw-5 |
| 5 | - - | T︰T | ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 6 | ++ | A︰A | Ty-1/Ty-1 | + + | G︰G | Mi-1/Mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 7 | ++ | A︰A | Ty-1/Ty-1 | + + | G︰G | Mi-1/Mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 8 | ++ | A︰A | Ty-1/Ty-1 | + + | G︰G | Mi-1/Mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 9 | + + | A︰A | Ty-1/Ty-1 | + + | G︰G | Mi-1/Mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 10 | + + | A︰A | Ty-1/Ty-1 | + + | G︰G | Mi-1/Mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 11 | - - | T︰T | ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 12 | - - | T︰T | ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 13 | - - | T︰T | ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 14 | - - | T︰T | ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 15 | - - | T︰T | ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 16 | - - | T︰T | ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | + | T︰T | Tm-22/Tm-22 | + + | A︰A | Frl/Frl | -- | A︰A | sw-5/sw-5 |
| 17 | - - | T︰T | ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | + | T︰T | Tm-22/Tm-22 | + + | A︰A | Frl/Frl | -- | A︰A | sw-5/sw-5 |
| 18 | + - | A︰T | Ty-1/ty-1 | +- | G︰A | Mi-1/mi-1 | + | T︰C | Tm-22/tm-22 | + - | A︰T | Frl/frl | -- | A︰A | sw-5/sw-5 |
| 19 | + - | A︰T | Ty-1/ty-1 | + + | G︰G | Mi-1/Mi-1 | + | T︰C | Tm-22/tm-22 | + - | A︰T | Frl/frl | -- | A︰A | sw-5/sw-5 |
| 20 | + - | A︰T | Ty-1/ty-1 | +- | G︰A | Mi-1/mi-1 | + | T︰C | Tm-22/tm-22 | + - | A︰T | Frl/frl | -- | A︰A | sw-5/sw-5 |
| 21 | + - | A︰T | Ty-1/ty-1 | +- | G︰A | Mi-1/mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 22 | - - | T︰T | ty-1/ty-1 | +- | G︰A | Mi-1/mi-1 | + | T︰T | Tm-22/Tm-22 | + + | A︰A | Frl/Frl | -- | A︰A | sw-5/sw-5 |
| 23 | + + | A︰A | Ty-1/Ty-1 | -- | A︰A | mi-1/mi-1 | + | T︰T | Tm-22/Tm-22 | + + | A︰A | Frl/Frl | -- | A︰A | sw-5/sw-5 |
| 24 | + + | A︰A | Ty-1/Ty-1 | -- | A︰A | mi-1/mi-1 | + | T︰T | Tm-22/Tm-22 | + + | A︰A | Frl/Frl | -- | A︰A | sw-5/sw-5 |
| 25 | + - | A︰T | Ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | + | T︰C | Tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 26 | + - | A︰T | Ty-1/ty-1 | +- | G︰A | Mi-1/mi-1 | + | T︰C | Tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 27 | + - | A︰T | Ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | - - | C︰C | tm-22/tm-22 | + + | A︰A | Frl/Frl | -- | A︰A | sw-5/sw-5 |
| 28 | + - | A︰T | Ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | - - | C︰C | tm-22/tm-22 | + + | A︰A | Frl/Frl | -- | A︰A | sw-5/sw-5 |
| 29 | + + | A︰A | Ty-1/Ty-1 | + + | G︰G | Mi-1/Mi-1 | + | T︰T | Tm-22/Tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 30 | + + | A︰A | Ty-1/Ty-1 | + + | G︰G | Mi-1/Mi-1 | + | T︰T | Tm-22/Tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 31 | + + | A︰A | Ty-1/Ty-1 | + + | G︰G | Mi-1/Mi-1 | + | T︰T | Tm-22/Tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 32 | + + | A︰A | Ty-1/Ty-1 | + + | G︰G | Mi-1/Mi-1 | + | T︰T | Tm-22/Tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 33 | + + | A︰A | Ty-1/Ty-1 | + + | G︰G | Mi-1/Mi-1 | + | T︰T | Tm-22/Tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 34 | + + | A︰A | Ty-1/Ty-1 | + + | G︰G | Mi-1/Mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 35 | + + | A︰A | Ty-1/Ty-1 | + + | G︰G | Mi-1/Mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 36 | + + | A︰A | Ty-1/Ty-1 | + + | G︰G | Mi-1/Mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 37 | - - | T︰T | ty-1/ty-1 | + + | G︰G | Mi-1/Mi-1 | + | T︰T | Tm-22/Tm-22 | + + | A︰A | Frl/Frl | -- | A︰A | sw-5/sw-5 |
| 38 | - - | T︰T | ty-1/ty-1 | + + | G︰G | Mi-1/Mi-1 | + | T︰T | Tm-22/Tm-22 | + + | A︰A | Frl/Frl | -- | A︰A | sw-5/sw-5 |
| 39 | - - | T︰T | ty-1/ty-1 | + + | G︰G | Mi-1/Mi-1 | + | T︰T | Tm-22/Tm-22 | + + | A︰A | Frl/Frl | -- | A︰A | sw-5/sw-5 |
| 40 | - - | T︰T | ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | + | T︰T | Tm-22/Tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 41 | - - | T︰T | ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | + | T︰T | Tm-22/Tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 42 | + + | A︰A | Ty-1/Ty-1 | -- | A︰A | mi-1/mi-1 | + | T︰T | Tm-22/Tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 43 | + + | A︰A | Ty-1/Ty-1 | -- | A︰A | mi-1/mi-1 | + | T︰T | Tm-22/Tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 44 | + + | A︰A | Ty-1/Ty-1 | -- | A︰A | mi-1/mi-1 | + | T︰T | Tm-22/Tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 45 | + + | A︰A | Ty-1/Ty-1 | -- | A︰A | mi-1/mi-1 | + | T︰T | Tm-22/Tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 46 | + + | A︰A | Ty-1/Ty-1 | -- | A︰A | mi-1/mi-1 | + | T︰T | Tm-22/Tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 47 | - - | T︰T | ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| 48 | - - | T︰T | ty-1/ty-1 | -- | A︰A | mi-1/mi-1 | - - | C︰C | tm-22/tm-22 | - - | T︰T | frl/frl | -- | A︰A | sw-5/sw-5 |
| [1] |
Allen A M, Barker G L, Berry S T, Coghill J A, Gwilliam R, Kirby S, Robinson P, Brenchley R C, D’Amore R, McKenzie N, Waite D, Hall A, Bevan M, Hall N, Edwards K J. 2011. Transcript-specific,single-nucleotide polymorphism discovery and linkage analysis in hexaploid bread wheat(Triticum aestivum L.). Plant Biotechnology Journal, 9 (9):1086-1099.
doi: 10.1111/pbi.2011.9.issue-9 URL |
| [2] |
Arens P, Mansilla C, Deinum D, Cavellini L, Moretti A, Rolland S, Van D S, Calvache D, Ponz F, Collonnier C, Mathis R, Smilde D, Caranta C, Vosman B. 2011. Development and evaluation of robust molecular markers linked to disease resistance in tomato for distinctness,uniformity and stability testing. Theoretical and Applied Genetics, 120 (3):655-664.
doi: 10.1007/s00122-009-1183-2 URL |
| [3] |
Berhanu T E, Veronica O, Mosisa W, Biswanath D, Michael O, Maryke L, Kassa S. 2015. Comparison of Kompetitive Allele Specific PCR(KASP)and genotyping by sequencing(GBS)for quality control analysis in maize. BMC Genomics, 16 (1):908-919.
doi: 10.1186/s12864-015-2180-2 URL |
| [4] |
Devran Z, Kahveci E. 2019. Development and validation of a user-friendly KASP marker for the Sw-5 locus in tomato. Australasian Plant Pathology, 48 (5):503-507.
doi: 10.1007/s13313-019-00651-1 URL |
| [5] |
Dianese R, Noronha F M, Goldbach R, Kormelink R, Inoue N A, Resende R, Boiteux L. 2010. Development of a locus-specific,co-dominant SCAR marker for assisted-selection of the Sw-5(Tospovirus resistance)gene cluster in a wide range of tomato accessions. Molecular Breeding, 25 (1):133-142.
doi: 10.1007/s11032-009-9313-8 URL |
| [6] | Ge Nai-peng, Cui Long, Li Han-xia, Ye Zhi-biao. 2014. Double SNP marker of Ty-1 alleles in a co-dominant manner in tomato. Acta Horticulturae Sinica, 41 (8):1583-1590. (in Chinese) |
| 葛乃蓬, 崔龙, 李汉霞, 叶志彪. 2014. 番茄抗黄化曲叶病毒基因Ty-1的双重SNP标记的开发. 园艺学报, 41 (8):1583-1590. | |
| [7] | Grogan S M, Brown-Guedira G, Haley S D, McMaster G S, Reid S D, Smith J, Byrne P F. 2016. Allelic variation in developmental genes and effects on winter wheat heading date in the U. S. great plains. PLoS ONE, 11:e0152852. |
| [8] | Guo Jia-jin, Zhang Sheng, Cheng Lin, Chen Fu-dong, Zhang Xiao-yan, Dong Tian, Li Xiao-jie, Liu Yue, Wang Ru-fang, Dong Jing, Yu Cai-yu. 2016. High-throughput molecular marker for identifying neck rot and root rot resistance of tomatoes and marking method and application thereof. China:CN105671190A, 2016-06-15. (in Chinese) |
| 国家进, 张生, 程琳, 陈福东, 张晓燕, 董甜, 李晓杰, 刘岳, 王如芳, 董静, 于彩玉. 2016. 鉴定番茄颈腐根腐病抗性的高通量分子标记及其标记方法与应用. 中国:CN105671190A, 2016-06-15. | |
| [9] | He C, Holme J, Anthony J. 2014. SNP genotyping:the KASP assay. Methods in Molecular Biology, 1145:75-86. |
| [10] | Hu Xia, Wang Rong, Li Fei-fei, Zhou Tao, Yang Wen-cai. 2014. Development of new marker for tomato TYLCV resistant gene and its application in selection of multi resistant gene pyramiding. China Vegetables,(10):18-23. (in Chinese) |
| 胡霞, 王蓉, 李菲菲, 周涛, 杨文才. 2014. 番茄抗TYLCV基因新标记的开发及其在多抗聚合选择中的应用. 中国蔬菜,(10):18-23. | |
| [11] | Jia Yu-yan, Chen Hong. 2003. Molecular genetic marker of single nucleotide polymorphism and its application. China Cattle Science, 29 (1):42-45. (in Chinese) |
| 贾玉艳, 陈宏. 2003. SNP分子标记的研究及应用. 中国牛业科学, 29 (1):42-45. | |
| [12] | Kong Hui-li, Jing Li-jun, He Zong-shun, Zhang Long-yu, Xia Qin-fang, Zhang Wen-li, Sun Ji-qing, Wei Yi, Yu Hui-hui, Zhou Fa-song. 2018. Molecular marker for identifying resistance gene of tomato mosaic virus(TOMV)and application thereof. China:CN108330201A, 2018-07-27. (in Chinese) |
| 孔会利, 井立军, 何宗顺, 张龙雨, 夏琴芳, 张文丽, 孙吉庆, 韦懿, 喻辉辉, 周发松. 2018. 鉴定番茄花叶病毒抗性基因的分子标记及其应用. 中国:CN108330201A, 2018-07-27. | |
| [13] | Kuang Meng, Wang Yan-qin, Zhou Da-yun, Ma Lei, Fang Dan, Xu Shuang-jiao, Yang Wei-hua, Wei Shou-jun, Ma Zhi-ying. 2016. High-throughput genotyping assay technology for cotton hybrid purity based on single-copy SNP markers. Cotton Science, 28 (3):227-233. (in Chinese) |
| 匡猛, 王延琴, 周大云, 马磊, 方丹, 徐双娇, 杨伟华, 魏守军, 马峙英. 2016. 基于单拷贝SNP标记的棉花杂交种纯度高通量检测技术. 棉花学报, 28 (3):227-233. | |
| [14] | Li Yong-ping, Kang Jian-ban, Lin Hui. 2016. Optimization and verification of ISSR-PCR reaction system for Lycopersicon esculentum. Journal of Agriculture, 6 (3):52-56. (in Chinese) |
| 李永平, 康建坂, 林珲. 2016. 番茄ISSR-PCR反应体系的优化与验证. 农学学报, 6 (3):52-56. | |
| [15] | Li Zong-jun, Wang Xian-yu, Liu Meng-jiao, Cui Xin-yue, Zhao Xiong, Chen Peng, Ou Qing-qing. 2019. Auxiliary screening of polymeric multi-resistance tomato materials using KASP molecular marker technology. China Vegetables,(8):42-46. (in Chinese) |
| 李宗俊, 王先裕, 刘梦姣, 崔馨月, 赵雄, 陈鹏, 欧青青. 2019. 利用KASP分子标记技术辅助筛选多抗番茄材料. 中国蔬菜,(8):42-46. | |
| [16] | Liang Shuang, Yuan Yu-xiang, Zhang Xiao-wei, Xue Yin-ge, Yao Qiu-ju, Jiang Wu-sheng, Tian Bao-ming, Zhang Qiang, Zhao Yan-yan. 2012. Optimization of InDel-PCR reaction system for Chinese cabbage(Brassica rapa L.ssp.pekinensis). Journal of Henan Agricultural Sciences, 41 (9):110-113. (in Chinese) |
| 梁爽, 原玉香, 张晓伟, 薛银鸽, 姚秋菊, 蒋武生, 田保明, 张强, 赵艳艳. 2012. 大白菜InDel-PCR反应体系的优化. 河南农业科学, 41 (9):110-113. | |
| [17] | Liu Lei, Deng Xuebin, Feng Jinging, Wang Jing, Sun Xiaorong, Shu Jinshuai, Li Junming. 2018. Exploring variant sites associated with Ve-1,I-2,Mi-1 and Pto genes in processing tomato by genome-wide association analysis. Acta Horticulturae Sinica, 45 (6):1089-1100. (in Chinese) |
| 刘磊, 邓学斌, 冯晶晶, 王静, 孙晓荣, 舒金帅, 李君明. 2018. 加工番茄Ve-1、I-2、Mi-1和Pto基因关联变异位点的挖掘. 园艺学报, 45 (6):1089-1100. | |
| [18] | Liu Li-hua, Pang Bin-shuang, Liu Yang-na, Li Hong-bo, Wang Na, Wang Zheng, Zhao Chang-ping. 2018. High-throughput identification mode for wheat varieties based on SNP markers. Journal of Triticeae Crops, 38 (5):529-534. (in Chinese) |
| 刘丽华, 庞斌双, 刘阳娜, 李宏博, 王娜, 王拯, 赵昌平. 2018. 基于SNP标记的小麦高通量身份鉴定模式. 麦类作物学报, 38 (5):529-534. | |
| [19] |
Liu Z Y, Zhu C S, Jiang Y, Tian Y L, Yu J, An H Z, Tang W J, Sun J, Tang J P, Chen G M, Zhai H Q, Wang C M, Wan J M. 2016. Association mapping and genetic dissection of nitrogen use efficiency- related traits in rice (Oryza sativa L.). Funct Integr Genomics, 16 (3):323-333.
doi: 10.1007/s10142-016-0486-z URL |
| [20] |
Lou H J, Dong L L, Zhang K P, Wang D W, Zhao M L, Li Y W, Rong C W, Qin H J, Zhang A M, Dong Z Y, Wang D W. 2017. High-throughput mining of e-genome-specific SNPs for characterizing thinopyrum elongatum introgressions in common wheat. Molecular Ecology Resources, 17 (6):1318-1329.
doi: 10.1111/men.2017.17.issue-6 URL |
| [21] |
Nair S K, Babu R, Magorokosho C, Mahuku G, Semagn K, Beyene Y, Das B, Makumbi D, Kumar P L, Olsen M. 2015. Fine mapping of Msv1,a major QTL for resistance to maize streak virus leads to development of production markers for breeding pipelines. Theor Appl Genet, 128:1839-1854.
doi: 10.1007/s00122-015-2551-8 URL |
| [22] |
Paux E, Sourdille P, Mackay I, Feuillet C. 2012. Sequence-based marker development in wheat:advances and applications to breeding. Biotechnology Advances, 30 (5):1071-1088.
doi: 10.1016/j.biotechadv.2011.09.015 URL |
| [23] |
Rasheed A, Wen W E, Gao F M, Zhai S N, Jin H, Liu J D, Guo Q, Zhang Y J, Dreisigacker S, Xia X C, He Z H. 2016. Development and validation of KASP assays for genes underpinning key economic traits in bread wheat. Theoretical and Applied Genetics, 129 (10):1843-1860.
doi: 10.1007/s00122-016-2743-x pmid: 27306516 |
| [24] |
Semagn K, Babu R, Hearne S, Olsen M. 2014. Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR(KASP):overview of the technology and its application in crop improvement. Molecular Breeding, 33 (1):1-14.
doi: 10.1007/s11032-013-9917-x URL |
| [25] |
Shi Z, Liu S M, James N, Prakash A, Khalid M, Li Z L. 2015. SNP identification and marker assay development for high-throughput selection of soybean cyst nematode resistance. BMC Genomics, 16 (1):314.
doi: 10.1186/s12864-015-1531-3 URL |
| [26] | Song Jian-jun, Wang Lin-shan, Tian Peng, Wang Rui-fei. 2012. Research progress on molecular marker for disease resistance genes in tomato. Journal of Northeast Agricultural University, 43 (4):1-10. (in Chinese) |
| 宋建军, 王琳珊, 田鹏, 王瑞飞. 2012. 番茄主要病害抗病基因分子标记的研究进展. 东北农业大学学报, 43 (4):1-10. | |
| [27] |
Staniaszek M, Szczechura W, Marczewski W. 2014. Identification of a new molecular marker C2- 25 linked to the Fusarium oxysporum f.sp radicis-lycopersici resistance Frl gene in tomato. Czech Journal of Genetics and Plant Breeding, 50 (4):285-287.
doi: 10.17221/CJGPB URL |
| [28] |
Steele K, Quinton M, Amgai R, Dhakal R, Khatiwada S, Vyas D, Heine M, Witcombe J. 2018. Accelerating public sector rice breeding with high-density KASP markers derived from whole genome sequencing of indica rice. Molecular Breeding, 38:38.
doi: 10.1007/s11032-018-0777-2 URL |
| [29] |
Su X M, Zhu G T, Huang Z J, Wang X X, Guo Y M, Li B J, Du Y C, Yang W C, Gao J C. 2019. Fine mapping and molecular marker development of the Sm gene conferring resistance to gray leaf spot(Stemphylium spp.)in tomato. Theoretical and Applied Genetics, 132 (4):871-882.
doi: 10.1007/s00122-018-3242-z URL |
| [30] |
Tan C T, Yu H J, Yang Y, Xu X Y, Chen M S, Rudd J C, Xue Q W, Ibrahim A M H, Garza L, Wang S C, Liu S Y. 2017. Development and validation of KASP markers for the greenbug resistance gene Gb7 and the Hessian fly resistance gene H32in wheat. Theor Appl Genet, 130:1867-1884.
doi: 10.1007/s00122-017-2930-4 URL |
| [31] |
Trick M, Adamski N, Mugford S G, iang C C, Febrer M, Uauy C. 2012. Combining SNP discovery from next-generation sequencing data with bulked segregant analysis(BSA)to fine-map genes in polyploid wheat. BMC Plant Biology, 12:14.
doi: 10.1186/1471-2229-12-14 URL |
| [32] |
Wang D G, Fan J B, Siao C J, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, Kruglyak L, Stein L, Hsie L, Topaloglou T, Hubbell E, Robinson E, Mittmann M, Morris M S, Shen N, Kilburn D, Rioux J, Nusbaum C, Rozen S, Hudson T J, Lipshutz R, Chee M, Lander E S. 1998. Large-scale identification,mapping,and genotyping of single-nucleotide polymorphisms in the human genome. Science, 280 (5366):1077-1082.
pmid: 9582121 |
| [33] | Wang Hui, Wang Shuang, Jiang Tong, Yang Yan-jie, Lin Duo. 2018. Construction and application of InDel-PCR system of dry pepper. Jiangsu Agricultural Sciences, 46 (10):28-30. (in Chinese) |
| 王辉, 王爽, 姜童, 杨延杰, 林多. 2018. 干制辣椒InDel-PCR反应体系的构建及应用. 江苏农业科学, 46 (10):28-30. | |
| [34] |
Wen C L, Mao A J, Dong C J, Liu H Y, Yu S C, Guo Y D, Weng Y Q, Xu Y. 2015. Fine genetic mapping of target leaf spot resistance gene cca-3 in cucumber,Cucumis sativus L. Theor Appl Genet, 128 (12):2495-2506.
doi: 10.1007/s00122-015-2604-z URL |
| [35] |
Wu J H, Liu S J, Wang Q L, Zeng Q D, Mu J M, Huang S, Yu S Z, Han D J, Kang Z S. 2018. Rapid identification of an adult plant stripe rust resistance gene in hexaploid wheat by high-throughput SNP array genotyping of pooled extremes. Theoretical and Applied Genetics, 131 (1):43-58.
doi: 10.1007/s00122-017-2984-3 URL |
| [36] | Wu Yuan-yuan, Li Hai-tao, Zhang Zi-jun, Zou Qing-dao. 2010. Research progress in molecular marker of disease resistant genes in tomato. Guizhou Agricultural Sciences, 38 (2):27-31. (in Chinese) |
| 吴媛媛, 李海涛, 张子君, 邹庆道. 2010. 番茄抗病基因分子标记研究进展. 贵州农业科学, 38 (2):27-31. | |
| [37] | Yang Chao-sha, Wang Guo-hua, Wu Zhi-ming, Yin Qing-zhen, Yin Wei-ping. 2018. Detection of resistant genes to tomato yellow leaf curl virus disease and root-knot nematode in tomato. Journal of Hebei Agricultural University, 41 (2):77-80. (in Chinese) |
| 杨超沙, 王国华, 吴志明, 尹庆珍, 尹伟平. 2018. 番茄黄化曲叶病毒及根结线虫抗性基因的检测. 河北农业大学学报, 41 (2):77-80. | |
| [38] | Yang Shuangjuan, Yuan Yuxiang, Wei Xiaochun, Wang Zhiyong, Zhao Yanyan, Yao Qiuju, Zhang Xiaowei. 2018. Optimization and establishment of KASP reaction system for Chinese cabbage(Brassica rapa L. ssp. Pekinensis). Acta Horticulturae Sinica, 48 (12):2442-2452. (in Chinese) |
| 杨双娟, 原玉香, 魏小春, 王志勇, 赵艳艳, 姚秋菊, 张晓伟. 2018. 大白菜KASP反应体系的优化与建立. 园艺学报, 48 (12):2442-2452. | |
| [39] | Yang Shuang-juan, Wang Zhi-Yong, Zhao Yan-yan, Wei Xiao-chun, Yuan Yu-xiang, Zhang Xiao-wei. 2020. Development of KASP marker for bolting related gene BrFLC1 in Chinese cabbage(Brassica rapa L.ssppekinensis). Journal of Nuclear Agricultural Sciences, 34 (2):265-272. (in Chinese) |
| 杨双娟, 王志勇, 赵艳艳, 魏小春, 原玉香, 张晓伟. 2020. 大白菜抽薹相关基因BrFLC1的KASP标记开发. 核农学报, 34 (2):265-272. | |
| [40] | Yuan Yu-xiang, Sun Ri-fei, Zhang Xiao-wei, Wu Jian, Xu Dong-hui, Zhang Hui, He Jiang-ming, Zhang Yan-guo, Wang Xiao-wu. 2008. A CAPS marker linked to bolting related gene BrFLC1 in Brassica rapa. Acta Horticulturae Sinica, 35 (11):1635-1640. (in Chinese) |
| 原玉香, 孙日飞, 张晓伟, 武剑, 徐东辉, 张慧, 和江明, 张延国, 王晓武. 2008. 芸薹种作物抽薹相关基因BrFLC1的CAPS标记. 园艺学报, 35 (11):1635-1640. | |
| [41] | Zhang Qiang, Zhang Tao, Chang Xiao-ke, Han Ya-nan, Cheng Zhi-fang, Liu Wei, Wang Bin, Yao Qiu-ju. 2019. Transformation and application of one molecular marker from SCAR to KASP in pepper CMS. cta Agriculturae Boreali-Sinica., 34 (5):93-98. (in Chinese) |
| 张强, 张涛, 常晓轲, 韩娅楠, 程志芳, 刘卫, 王彬, 姚秋菊. 2019. 一个辣椒胞质雄性不育SCAR标记的KASP转化及其应用. 华北农学报, 34 (5):93-98. | |
| [42] | Zhao Yong, Liu Xiao-dong, Zhao Hong-kun, Yuan Cui-ping, Qi Guang-xun, Wang Yu-min, Dong Ying-shan. 2017. Comparison of methods for SNP genotyping in soybean. Molecular Plant Breeding, 15 (9):3540-3546. (in Chinese) |
| 赵勇, 刘晓冬, 赵洪锟, 袁翠平, 齐广勋, 王玉民, 董英山. 2017大豆SNP分型方法的比较. 分子植物育种, 15 (9):3540-3546. | |
| [43] | Zhu Ming-tao, Sun Ya-lin, Zheng Sha, Zhang Xiao-li, Wang Tao-tao, Ye Zhi-biao, Li Han-xia. 2010. Pyramiding disease resistance genes by molecular marker-assisted selection in tomato. Acta Horticulturae Sinica, 37 (9):1416-1422. (in Chinese) |
| 朱明涛, 孙亚林, 郑莎, 张晓黎, 王涛涛, 叶志彪, 李汉霞. 2010. 分子标记辅助聚合番茄抗病基因育种. 园艺学报, 37 (9):1416-1422. | |
| [44] |
Zübeyir D, Atilla G, Lütfiye M. 2016. Development of molecular markers for the Mi-1 gene in tomato using the KASP genotyping assay. Horticulture Environment and Biotechnology, 57 (2):156-160.
doi: 10.1007/s13580-016-0028-6 URL |
| [1] | REN Hailong, XU Donglin, ZHANG Jing, ZOU Jiwen, LI Guangguang, ZHOU Xianyu, XIAO Wanyu, and SUN Yijia. Establishment of SNP Fingerprinting and Identification of Chinese Flowering Cabbage Varieties Based on KASP Genotyping [J]. Acta Horticulturae Sinica, 2023, 50(2): 307-318. |
| [2] | SHI Hongli, LI La, GUO Cuimei, YU Tingting, JIAN Wei, YANG Xingyong. Isolation,Identification and Analysis of Biocontrol Ability of Biocontrol Strain TL1 Against Tomato Botrytis cinerea [J]. Acta Horticulturae Sinica, 2023, 50(1): 79-90. |
| [3] | HU Jingyu, QUE Kaijuan, MIAO Tianli, WU Shaozheng, WANG Tiantian, ZHANG Lei, DONG Xian, JI Pengzhang, DONG Jiahong. Identification of Tomato Spotted Wilt Orthotospovirus Infecting Iris tectorum [J]. Acta Horticulturae Sinica, 2023, 50(1): 170-176. |
| [4] | ZHENG Jirong, WANG Tonglin, and HU Songshen. A New Tomato Cultivar‘Hangza 603’with High Quality [J]. Acta Horticulturae Sinica, 2022, 49(S2): 103-104. |
| [5] | ZHENG Jirong and WANG Tonglin. A New Tomato Cultivar‘Hangza 601’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 105-106. |
| [6] | ZHENG Jirong and WANG Tonglin. A New Cherry Tomato Cultivar‘Hangza 503’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 107-108. |
| [7] | HUANG Tingting, LIU Shuqin, ZHANG Yongzhi, LI Ping, ZHANG Zhihuan, and SONG Libo. A New Cherry Tomato Cultivar‘Yingshahong 4’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 109-110. |
| [8] | ZHANG Qianrong, LI Dazhong, QIU Boyin, LIN Hui, MA Huifei, YE Xinru, LIU Jianting, ZHU Haisheng, and WEN Qingfang. A New Tomato Cultivar‘Minnongke 2’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 73-74. |
| [9] | HAN Shuai, WU Jie, ZHANG Heqing, XI Yadong. Identification and Sequence Analysis of Tomato Spotted Wilt Orthotospovirus Infecting Lettuce in Sichuan [J]. Acta Horticulturae Sinica, 2022, 49(9): 2007-2016. |
| [10] | WU Fan, JIANG Junhao, LU Shan, ZHANG Nan, QIU Shuai, WEI Jianfen, SHEN Baichun. Research Progress on Germplasm Resources and Utilization of Hydrangea in China [J]. Acta Horticulturae Sinica, 2022, 49(9): 2037-2050. |
| [11] | CHEN Lilang, YANG Tianzhang, CAI Ruping, LIN Xiaoman, DENG Nankang, CHE Haiyan, LIN Yating, KONG Xiangyi. Molecular Detection and Identification of Viruses from Passiflora edulis in Hainan [J]. Acta Horticulturae Sinica, 2022, 49(8): 1785-1794. |
| [12] | ZHANG Yue, SUO Yujing, SUN Peng, HAN Weijuan, DIAO Songfeng, LI Huawei, ZHANG Jiajia, FU Jianmin, LI Fangdong. Analysis on Fruit Morphological Diversity of Persimmon Germplasm Resources [J]. Acta Horticulturae Sinica, 2022, 49(7): 1473-1490. |
| [13] | LÜ Zhengxin, HE Yanqun, JIA Dongfeng, HUANG Chunhui, ZHONG Min, LIAO Guanglian, ZHU Yi, YUAN Kaichang, LIU Chuanhao, XU Xiaobiao. Genetic Diversity Analysis of Phenotypic Traits for Kiwifruit Germplasm Resources [J]. Acta Horticulturae Sinica, 2022, 49(7): 1571-1581. |
| [14] | LI Pingping, ZHANG Xiang, LIU Yuting, XIE Zhihe, ZHANG Ruihao, ZHAO Kai, LÜ Junheng, WANG Ziran, WEN Jinfen, ZOU Xuexiao, DENG Minghua. Studies on the Relationship Between Pigment Composition and Fruit Coloration of 63 Peppers [J]. Acta Horticulturae Sinica, 2022, 49(7): 1589-1601. |
| [15] | LU Tao, YU Hongjun, LI Qiang, JIANG Weijie. Effects of Leaf and Fruit Quantity Regulation on Growth,Fruit Quality and Yield of Tomato [J]. Acta Horticulturae Sinica, 2022, 49(6): 1261-1274. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd