
Acta Horticulturae Sinica ›› 2021, Vol. 48 ›› Issue (10): 1847-1858.doi: 10.16420/j.issn.0513-353x.2021-0090
• Research Papers • Previous Articles Next Articles
LIU Yue1, LI Yueqing2, MENG Xiangyu3, HUANG Jing1, TANG Hao1, LI Yuanheng1, GAO Xiang2, ZHAO Chunli1,*( )
)
Received:2021-06-09
Revised:2021-08-30
Online:2021-10-25
Published:2021-11-01
Contact:
ZHAO Chunli
E-mail:zcl8368@163.com
CLC Number:
LIU Yue, LI Yueqing, MENG Xiangyu, HUANG Jing, TANG Hao, LI Yuanheng, GAO Xiang, ZHAO Chunli. Cloning and Functional Characterization of Chalcone Synthase Genes(CmCHS)from Clivia miniata[J]. Acta Horticulturae Sinica, 2021, 48(10): 1847-1858.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2021-0090
| 用途 Use | 基因 Gene | 引物序列(5′-3′) Primer sequence |
|---|---|---|
| 基因ORF的克隆 Cloning the full-length of ORF | CmCHS1 | F:ATGGTGAGTATCGATGAGATT;R:CATCGCGCCGATGATTAATT |
| CmCHS2 | F:ATGGTGAGTGTCCATGAGATTCGC;R:CACGATGTGTCGCATCGATAAATT | |
| CmCHS3 | F:ATGGTGAGCGTGGAAGAGATC;R:GGTATCACACCGATGATTAATT | |
| CmCHS4 | F:ATGGCCACAATTGAGGAGATCAG;R:ACGCTTCACACCGCTAGTTAACT | |
| T7 | F:TAATACGACTCACTATAGGG;R:GCTAGTTATTGCTCAGCGG | |
| 实时荧光定PCR Quantitative Real-time PCR | CmCHS1 | F:GACATGCCCGGTGCAGACTACCA;R:AGCCGTGATCTCCGAGCAGACG |
| CmCHS2 | F:GACGAGACAAGTGTTGAGCGAGTA;R:TCCAGGTCCGAATCCAAAGAGC | |
| CmCHS3 | F:AGGAGCGATAGACGGGCACTTA;R:GAACAGCGAGTTCCAATCTAACATC | |
| CmCHS4 | F:GGAGGTGCTGAAGGAGTATGGGAACA;R:TCCAGGGCCGAACCCAAATAGA | |
| CmActin | F:GCATCACACCTTCTACAA;R:CATTGTAGAAGGTGTGATG | |
| 制备重组蛋白 Preparation of recombinant protein | CmCHS1 | F:TGGCTGATATCGGATCCATGGTGAGTATCGATGAG; R:CGACGGAGCTCGAATTCTTAATTAGTAGCCGCGCT |
| CmCHS2 | F:TGGCTGATATCGGATCCATGGTGAGTGTCCATGAG; R:CGACGGAGCTCGAATTCTTAAATAGCTACGCTGTG | |
| CmCHS3 | F:TGGCTGATATCGGATCCATGGTGAGCGTGGAAGAG; R:CGACGGAGCTCGAATTCTTAATTAGTAGCCACACT | |
| CmCHS4 | F:TGGCTGATATCGGATCCATGGCCACAATTGAGGAG; R:CGACGGAGCTCGAATTCTCAATTGATCGCCACACT | |
| 亚细胞定位 Subcellular | GFP(35S:GFP) | F:CTGATTACGCTCATATGATGGTGAGCAAGGGCGAG; R:AGGATTCAATCTTAAGTTACTTGTACAGCTCGTCCATG |
| localization | (GFP)CmCHS1 | F:GACGAGCTGTACAAGCATATGGTGAGTATCGAT; R:CAACAGGATTCAATCTTAAGTTAATTAGTAGCCGC |
| (GFP)CmCHS2 | F:GACGAGCTGTACAAGCATATGGTGAGTGTCCAT; R:CAACAGGATTCAATCTTAAGTTAAATAGCTACGCT | |
| (GFP)CmCHS3 | F:GACGAGCTGTACAAGCATATGGTGAGCGTGGAA; R:CAACAGGATTCAATCTTAAGTTAATTAGTAGCCAC | |
| (GFP)CmCHS4 | F:GACGAGCTGTACAAGCATATGGCCACAATTGAG; R:CAACAGGATTCAATCTTAAGTCAATTGATCGCCAC |
Table 1 Primer sequences in this study
| 用途 Use | 基因 Gene | 引物序列(5′-3′) Primer sequence |
|---|---|---|
| 基因ORF的克隆 Cloning the full-length of ORF | CmCHS1 | F:ATGGTGAGTATCGATGAGATT;R:CATCGCGCCGATGATTAATT |
| CmCHS2 | F:ATGGTGAGTGTCCATGAGATTCGC;R:CACGATGTGTCGCATCGATAAATT | |
| CmCHS3 | F:ATGGTGAGCGTGGAAGAGATC;R:GGTATCACACCGATGATTAATT | |
| CmCHS4 | F:ATGGCCACAATTGAGGAGATCAG;R:ACGCTTCACACCGCTAGTTAACT | |
| T7 | F:TAATACGACTCACTATAGGG;R:GCTAGTTATTGCTCAGCGG | |
| 实时荧光定PCR Quantitative Real-time PCR | CmCHS1 | F:GACATGCCCGGTGCAGACTACCA;R:AGCCGTGATCTCCGAGCAGACG |
| CmCHS2 | F:GACGAGACAAGTGTTGAGCGAGTA;R:TCCAGGTCCGAATCCAAAGAGC | |
| CmCHS3 | F:AGGAGCGATAGACGGGCACTTA;R:GAACAGCGAGTTCCAATCTAACATC | |
| CmCHS4 | F:GGAGGTGCTGAAGGAGTATGGGAACA;R:TCCAGGGCCGAACCCAAATAGA | |
| CmActin | F:GCATCACACCTTCTACAA;R:CATTGTAGAAGGTGTGATG | |
| 制备重组蛋白 Preparation of recombinant protein | CmCHS1 | F:TGGCTGATATCGGATCCATGGTGAGTATCGATGAG; R:CGACGGAGCTCGAATTCTTAATTAGTAGCCGCGCT |
| CmCHS2 | F:TGGCTGATATCGGATCCATGGTGAGTGTCCATGAG; R:CGACGGAGCTCGAATTCTTAAATAGCTACGCTGTG | |
| CmCHS3 | F:TGGCTGATATCGGATCCATGGTGAGCGTGGAAGAG; R:CGACGGAGCTCGAATTCTTAATTAGTAGCCACACT | |
| CmCHS4 | F:TGGCTGATATCGGATCCATGGCCACAATTGAGGAG; R:CGACGGAGCTCGAATTCTCAATTGATCGCCACACT | |
| 亚细胞定位 Subcellular | GFP(35S:GFP) | F:CTGATTACGCTCATATGATGGTGAGCAAGGGCGAG; R:AGGATTCAATCTTAAGTTACTTGTACAGCTCGTCCATG |
| localization | (GFP)CmCHS1 | F:GACGAGCTGTACAAGCATATGGTGAGTATCGAT; R:CAACAGGATTCAATCTTAAGTTAATTAGTAGCCGC |
| (GFP)CmCHS2 | F:GACGAGCTGTACAAGCATATGGTGAGTGTCCAT; R:CAACAGGATTCAATCTTAAGTTAAATAGCTACGCT | |
| (GFP)CmCHS3 | F:GACGAGCTGTACAAGCATATGGTGAGCGTGGAA; R:CAACAGGATTCAATCTTAAGTTAATTAGTAGCCAC | |
| (GFP)CmCHS4 | F:GACGAGCTGTACAAGCATATGGCCACAATTGAG; R:CAACAGGATTCAATCTTAAGTCAATTGATCGCCAC |

Fig. 1 Sequence alignment of CHSs from Clivia miniata and other species AtCHS:Arabidopsis thaliana,AAA32771.1;FhCHS:Freesia hybrida,JF732897;GbCHS:Ginkgo biloba,AAT68477.1;ZmCHS:Zea mays,CAA42763.1;MsCHS:Medicago sativa,P30074.1;VvCHS:Vitis vinifera,BAA31259.1;VvSTS:Vitis vinifera,ABV82966.1.The yellow box represents three conserved catalytic residues in CHS. The green frame amino acid determines the specificity of CHS substrate;The red frame amino acid is the binding region of malonyl coenzyme A. The black rectangular box indicates the highly conserved domains of CHS. The blue circles represent important residues in binding to coumarinyl coenzyme A and residues specific to the cyclic reaction of CHS.
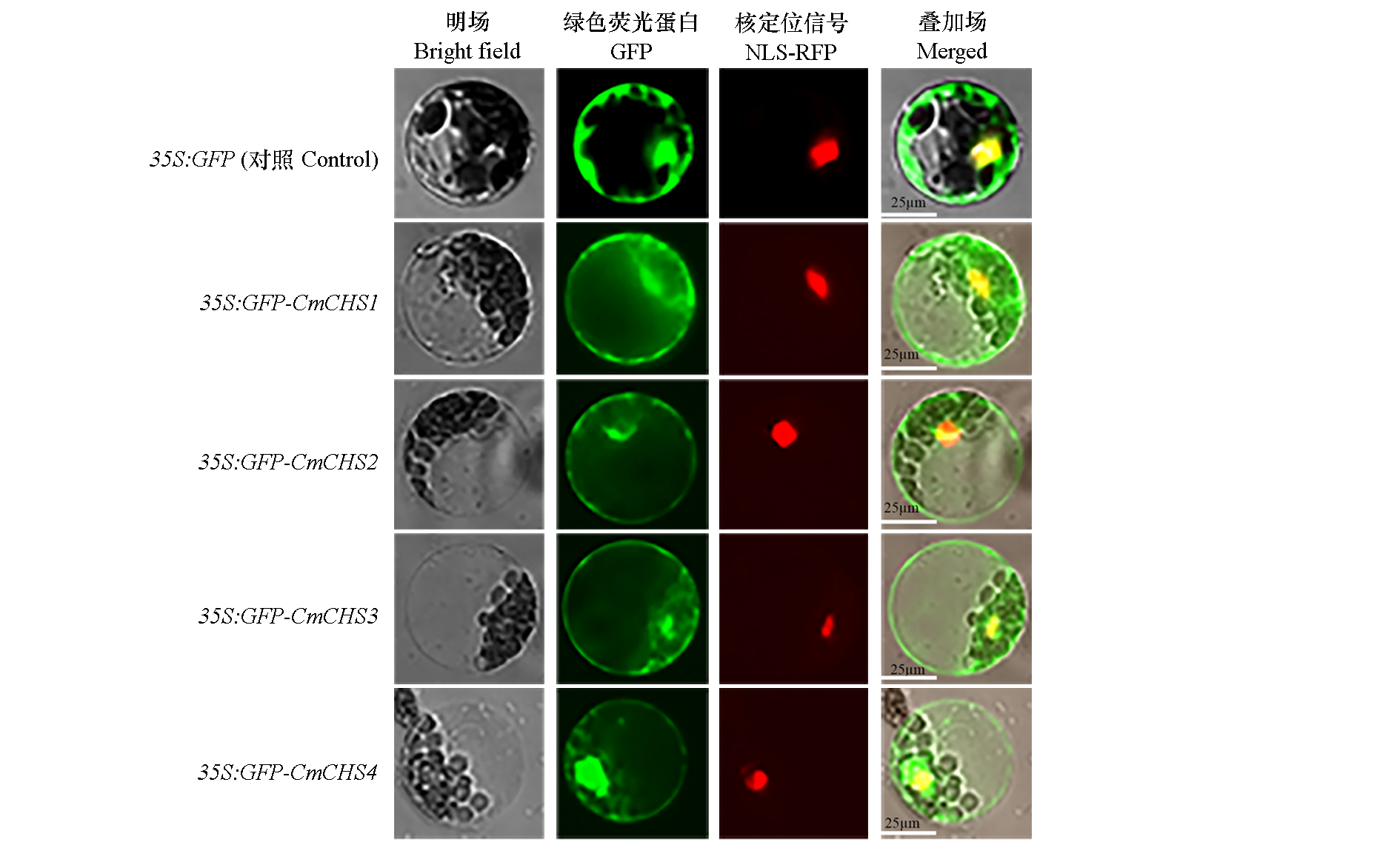
Fig. 3 Subcellular localization of CmCHSs Four recombinant plasmids were transiently transfected into wild-type Arabidopsis protoplasts,and incubated for 21 h at room temperature in the dark. Laser scanning confocal microscopy captured the fluorescent proteins.
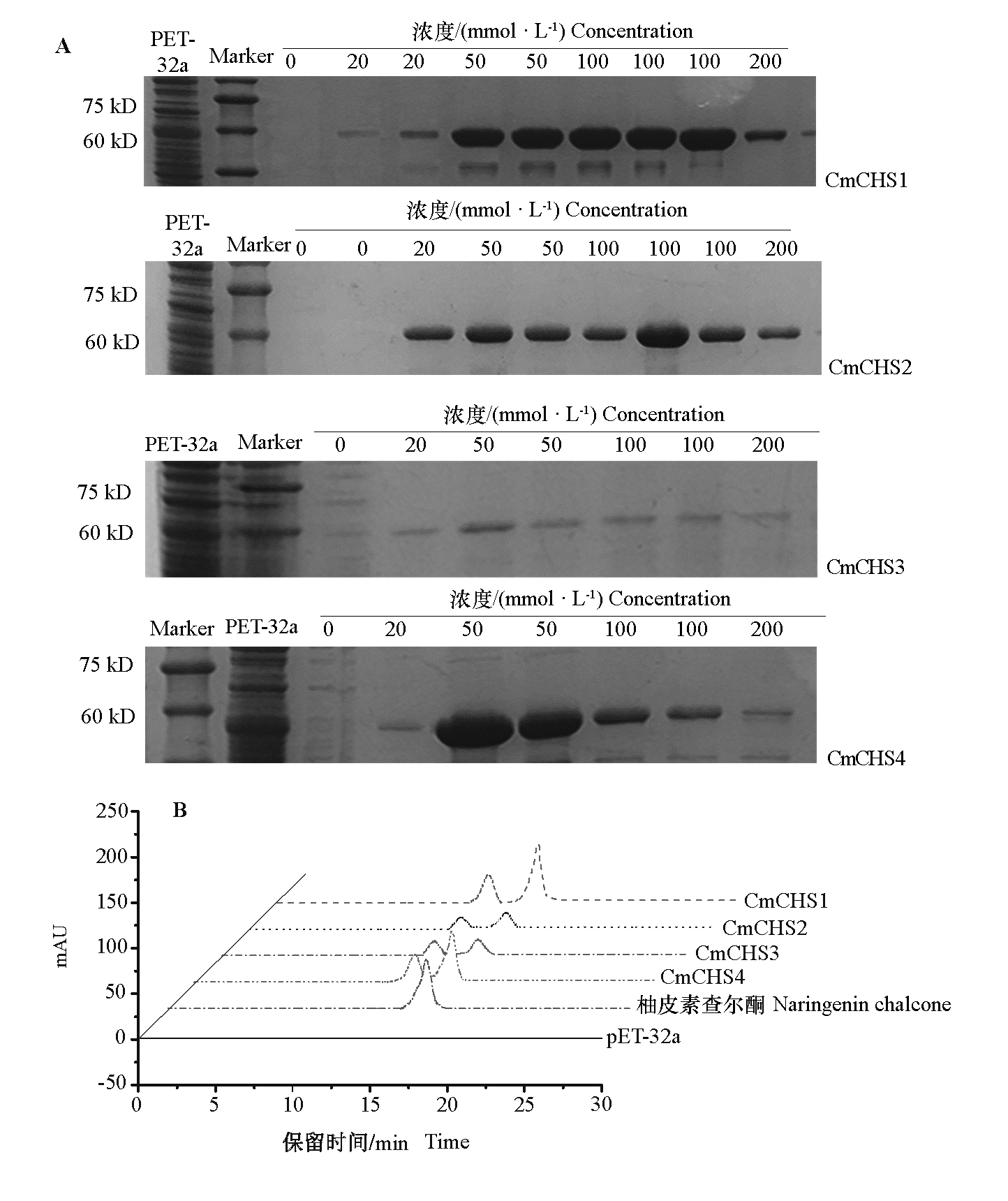
Fig. 5 In vitro enzymatic analysis of CmCHS1,CmCHS2,CmCHS3 and CmCHS4 A:The recombinant protein was extracted from sodium dodecyl sulfate polyacrylamide gel electrophoresis(SDS-PAGE);B:HPLC.
| [1] |
Abe I, Morita H. 2010. Structure and function of the chalcone synthase superfamily of plant type Ⅲ polyketide synthases. Natural Product Reports, 27 (6):809-838.
doi: 10.1039/b909988n URL |
| [2] |
Abe I, Takahashi Y, Noguchi H. 2002. Enzymatic formation of an unnatural C(6)-C(5) aromatic polyketide by plant type Ⅲ polyketide synthases. Organic Letters, 4 (21):3623-3626.
doi: 10.1021/ol0201409 URL |
| [3] |
Abe I, Utsumi Y, Oguro S, Noguchi H. 2004. The first plant type Ⅲ polyketide synthase that catalyzes formation of aromatic heptaketide. FEBS Letters, 562 (1-3):171-176.
doi: 10.1016/S0014-5793(04)00230-3 URL |
| [4] |
Agati G, Tattini M. 2010. Multiple functional roles of flavonoids in photoprotection. The New Phytologist, 186 (4):786-793.
doi: 10.1111/nph.2010.186.issue-4 URL |
| [5] | Aron P M, Kennedy J A. 2008. Flavan-3-ols:nature,occurrence and biological activity. Molecular Nutrition & Food Research, 52 (1):79-104. |
| [6] |
Austin M B, Noel J P. 2003. The chalcone synthase superfamily of type III polyketide synthases. Natural Product Reports, 20 (1):79-110.
doi: 10.1039/b100917f URL |
| [7] |
Deng X, Bashandy H, Ainasoja M, Kontturi J, Pietiäinen M, Laitinen R A, Albert V A, Valkonen J P, Elomaa P, Teeri T H. 2014. Functional diversification of duplicated chalcone synthase genes in anthocyanin biosynthesis of Gerbera hybrida. The New Phytologist, 201 (4):1469-1483.
doi: 10.1111/nph.2014.201.issue-4 URL |
| [8] |
Ferrer J L, Jez J M, Bowman M E, Dixon R A, Noel J P. 1999. Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nature Structural Biology, 6 (8):775-784.
pmid: 10426957 |
| [9] |
Forkmann G, Martens S. 2001. Metabolic engineering and applications of flavonoids. Current Opinion in Biotechnology, 12 (2):155-160.
pmid: 11287230 |
| [10] |
Franken P, Niesbach-Klösgen U, Weydemann U, Maréchal-Drouard L, Saedler H, Wienand U. 1991. The duplicated chalcone synthase genes C2and Whp(white pollen)of Zea mays are independently regulated;evidence for translational control of Whp expression by the anthocyanin intensifying gene in. The EMBO Journal, 10 (9):2605-2612.
doi: 10.1002/embj.1991.10.issue-9 URL |
| [11] |
Grotewold E. 2006. The genetics and biochemistry of floral pigments. Annual Review of Plant Biology, 57:761-780.
pmid: 16669781 |
| [12] |
Han Y, Zhao W, Wang Z, Zhu J, Liu Q. 2014. Molecular evolution and sequence divergence of plant chalcone synthase and chalcone synthase-like genes. Genetica, 142 (3):215-225.
doi: 10.1007/s10709-014-9768-3 URL |
| [13] |
Hermanns A S, Zhou X S, Xu Q, Tadmor Y, Li L. 2020. Carotenoid Pigment Accumulation in Horticultural Plants. Horticultural Plant Journal, 6 (6):343-360.
doi: 10.1016/j.hpj.2020.10.002 URL |
| [14] |
Jiang C, Schommer C K, Kim S Y, Suh D Y. 2006. Cloning and characterization of chalcone synthase from the moss,Physcomitrella patens. Phytochemistry, 67 (23):2531-2540.
doi: 10.1016/j.phytochem.2006.09.030 URL |
| [15] |
Joachim S. 1997. A family of plant-specific polyketide synthases:facts and predictions. Trends in Plant Science, 2 (10):373-378.
doi: 10.1016/S1360-1385(97)87121-X URL |
| [16] |
Kim S S, Grienenberger E, Lallemand B, Colpitts C C, Kim S Y, Souza C, Geoffroy P, Heintz D, Krahn D, Kaiser M, Kombrink E, Heitz T, Suh D Y, Legrand M, Douglas C J. 2010. Lap6/polyketide synthase a and lap5/polyketide synthase b encode hydroxyalkyl α-pyrone synthases required for pollen development and sporopollenin biosynthesis in Arabidopsis thaliana. The Plant Cell, 22 (12):4045-4066.
doi: 10.1105/tpc.110.080028 URL |
| [17] |
Koes R, Verweij W, Quattrocchio F. 2005. Flavonoids:a colorful model for the regulation and evolution of biochemical pathways. Trends in Plant Science, 10 (5):236-242.
doi: 10.1016/j.tplants.2005.03.002 URL |
| [18] |
Koes R E, Spelt C E, van den Elzen P J, Mol J N. 1989. Cloning and molecular characterization of the chalcone synthase multigene family of Petunia hybrida. Gene, 81 (2):245-257.
pmid: 2806915 |
| [19] |
Kong J M, Chia L S, Goh N K, Chia T F, Brouillard R. 2003. Analysis and biological activities of anthocyanins. Phytochemistry, 64 (5):923-933.
doi: 10.1016/S0031-9422(03)00438-2 URL |
| [20] |
Li Y, Shan X, Gao R, Han T, Zhang J, Wang Y, Kimani S, Wang L, Gao X. 2020. MYB repressors and MBW activation complex collaborate to fine-tune flower coloration in Freesia hybrida. Communications Biology, 3 (1):396.
doi: 10.1038/s42003-020-01134-6 URL |
| [21] |
Li Y, Shan X, Zhou L, Gao R, Yang S, Wang S, Wang L, Gao X. 2019. The R2R3-MYB Factor FhMYB 5 from Freesia hybrida contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis. Frontiers in Plant Science, 9:1935.
doi: 10.3389/fpls.2018.01935 URL |
| [22] |
Liou G, Chiang Y C, Wang Y, Weng J K. 2018. Mechanistic basis for the evolution of chalcone synthase catalytic cysteine reactivity in land plants. The Journal of Biological Chemistry, 293 (48):18601-18612.
doi: 10.1074/jbc.RA118.005695 URL |
| [23] |
Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCт method. Methods, 25 (4):402-408.
pmid: 11846609 |
| [24] |
Luan Y, Fu X, Lu P, Grierson D, Xu C. 2020. Molecular mechanisms determining the differential accumulation of carotenoids in plant species and varieties. Critical Reviews in Plant Sciences, 39 (2):1-15.
doi: 10.1080/07352689.2020.1724433 URL |
| [25] |
Morita H, Takahashi Y, Noguchi H, Abe I. 2000. Enzymatic formation of unnatural aromatic polyketides by chalcone synthase. Biochemical and Biophysical Research Communications, 279 (1):190-195.
pmid: 11112437 |
| [26] |
Morita Y, Saito R, Ban Y, Tanikawa N, Kuchitsu K, Ando T, Yoshikawa M, Habu Y, Ozeki Y, Nakayama M. 2012. Tandemly arranged chalcone synthase A genes contribute to the spatially regulated expression of siRNA and the natural bicolor floral phenotype in Petunia hybrida. The Plant Journal:for Cell and Molecular Biology, 70 (5):739-749.
doi: 10.1111/j.1365-313X.2012.04908.x URL |
| [27] |
Nakatsuka A, Izumi Y, Yamagishi M. 2003. Spatial and temporal expression of chalcone synthase and dihydroflavonol 4-reductase genes in the Asiatic hybrid lily. Plant Science, 165 (4):759-767.
doi: 10.1016/S0168-9452(03)00254-1 URL |
| [28] |
Napoli C A, Fahy D, Wang H Y, Taylor L P. 1999. White anther:a petunia mutant that abolishes pollen flavonol accumulation,induces male sterility,and is complemented by a chalcone synthase transgene. Plant Physiology, 120 (2):615-622.
pmid: 10364414 |
| [29] | Petroni K, Tonelli C. 2011. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Science:an International Journal of Experimental Plant Biology, 181 (3):219-229. |
| [30] |
Saigo T, Wang T, Watanabe M, Tohge T. 2020. Diversity of anthocyanin and proanthocyanin biosynthesis in land plants. Current Opinion in Plant Biology, 55:93-99.
doi: 10.1016/j.pbi.2020.04.001 URL |
| [31] |
Saito K, Yonekura-Sakakibara K, Nakabayashi R, Higashi Y, Yamazaki M, Tohge T, Fernie A R. 2013. The flavonoid biosynthetic pathway in Arabidopsis:structural and genetic diversity. Plant Physiology and Biochemistry, 72:21-34.
doi: 10.1016/j.plaphy.2013.02.001 URL |
| [32] |
Shan X, Li Y, Yang S, Gao R, Zhou L, Bao T, Han T, Wang S, Gao X, Wang L. 2019. A functional homologue of Arabidopsis TTG1 from Freesia interacts with bHLH proteins to regulate anthocyanin and proanthocyanidin biosynthesis in both Freesia hybrida and Arabidopsis thaliana. Plant Physiology and Biochemistry, 141:60-72.
doi: 10.1016/j.plaphy.2019.05.015 URL |
| [33] |
Shan X, Li Y, Yang S, Yang Z, Qiu M, Gao R, Han T, Meng X, Xu Z, Wang L, Gao X. 2020. The spatio-temporal biosynthesis of floral flavonols is controlled by differential phylogenetic MYB regulators in Freesia hybrida. New Phytologist, 228 (6):1864-1879.
doi: 10.1111/nph.v228.6 URL |
| [34] |
Shirley B W, Kubasek W L, Storz G, Bruggemann E, Koornneef M, Ausubel F M, Goodman H M. 1995. Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. The Plant Journal:For Cell and Molecular Biology, 8 (5):659-671.
doi: 10.1046/j.1365-313X.1995.08050659.x URL |
| [35] |
Sun W, Meng X, Liang L, Jiang W, Huang Y, He J, Hu H, Almqvist J, Gao X, Wang L. 2015. Molecular and biochemical analysis of chalcone synthase from Freesia hybrid in flavonoid biosynthetic pathway. PLoS ONE, 10 (3):e0119054.
doi: 10.1371/journal.pone.0119054 URL |
| [36] |
Tanaka Y, Ohmiya A. 2008. Seeing is believing: engineering anthocyanin and carotenoid biosynthetic pathways. Current Opinion in Biotechnology, 19 (2):190-197.
doi: 10.1016/j.copbio.2008.02.015 URL |
| [37] |
Tanaka Y, Sasaki N, Ohmiya A. 2008. Biosynthesis of plant pigments:anthocyanins,betalains and carotenoids. The Plant Journal, 54 (4):733-749.
doi: 10.1111/j.1365-313X.2008.03447.x URL |
| [38] |
Timoneda A, Feng T, Sheehan H, Walker-Hale N, Pucker B, Lopez-Nieves S, Guo R, Brockington S. 2019. The evolution of betalain biosynthesis in Caryophyllales. New Phytologist, 224 (1):71-85.
doi: 10.1111/nph.15980 pmid: 31172524 |
| [39] |
Václav T, Milena M, Tomáš V, Bořivoj K, Pavel H, Ladislav H. 2014. Chalcone synthase expression and pigments deposition in wheat with purple and blue colored caryopsis. Journal of Cereal Science, 59 (10):48-55.
doi: 10.1016/j.jcs.2013.10.008 URL |
| [40] |
Wang X, Wang X, Hu Q, Dai X, Tian H, Zheng K, Wang X, Mao T, Chen J G, Wang S. 2015. Characterization of an activation-tagged mutant uncovers a role of GLABRA 2 in anthocyanin biosynthesis in Arabidopsis. The Plant Journal, 83 (2):300-311.
doi: 10.1111/tpj.12887 URL |
| [41] |
Wang Z, Yu Q, Shen W, El Mohtar C A, Zhao X, Gmitter F G Jr. 2018. Functional study of CHS gene family members in citrus revealed a novel CHS gene affecting the production of flavonoids. BMC Plant Biology, 18 (1):189.
doi: 10.1186/s12870-018-1418-y URL |
| [42] |
Weng J K, Chapple C. 2010. The origin and evolution of lignin biosynthesis. The New Phytologist, 187 (2):273-285.
doi: 10.1111/nph.2010.187.issue-2 URL |
| [43] |
Winkel-Shirley B. 2001. Flavonoid biosynthesis. a colorful model for genetics,biochemistry,cell biology,and biotechnology. Plant Physiology, 126 (2):485-493.
pmid: 11402179 |
| [44] |
Yamazaki Y, Suh D Y, Sitthithaworn W, Ishiguro K, Kobayashi Y, Shibuya M, Ebizuka Y, Sankawa U. 2001. Diverse chalcone synthase superfamily enzymes from the most primitive vascular plant,Psilotum nudum. Planta, 214 (1):75-84.
pmid: 11762173 |
| [45] |
Yonekura-Sakakibara K, Higashi Y, Nakabayashi R. 2019. The origin and evolution of plant flavonoid metabolism. Frontiers in Plant Science, 10:943.
doi: 10.3389/fpls.2019.00943 pmid: 31428108 |
| [46] | Zhang Bi-xian, Zhu Yan-ming, Lai Yong-cai, Hu Xiao-mei, Li Wei, Li Wan, Bi Ying-dong, Xiao Jia-lei, Zhang Li-li. 2012. Research progress of plant chalcone synthase(CHS)and its gene. Journal of Anhui Agricultural Sciences, 40 (20):10376-10379.(in Chinese) |
| 张必弦, 朱延明, 来永才, 胡小梅, 李炜, 李琬, 毕影东, 肖佳雷, 张俐俐. 2012. 植物查尔酮合酶(CHS)及其基因的研究进展. 安徽农业科学, 40 (20):10376-10379. | |
| [47] | Zheng Yi, Zhang Hui, Wang Qinmei, Gao Yue, Zhang Zhihong, Sun Yuxin. 2020. Complete chloroplast genome sequence of Clivia miniata and its characteristics. Acta Horticulturae Sinica, 47 (12):2439-2450.(in Chinese) |
| 郑祎, 张卉, 王钦美, 高悦, 张志宏, 孙玉新. 2020. 大花君子兰叶绿体基因组及其特征. 园艺学报, 47 (12):2439-2450. | |
| [48] | Zhou L, Zheng K, Wang X, Tian H, Wang X, Wang S. 2014. Control of trichome formation in Arabidopsis by poplar single-repeat R3 MYB transcription factors. Frontiers in Plant Science, 5:262. |
| [49] |
Zhu K J, Wu Q J, Huang Y, Ye J L, Xu Q, Deng X X. 2020. Genome-wide characterization of cis-acting elements in the promoters of key carotenoid pathway genes from the main species of genus citrus. Horticultural Plant Journal, 6 (6):385-395.
doi: 10.1016/j.hpj.2020.10.003 URL |
| [1] | YE Zimao, SHEN Wanxia, LIU Mengyu, WANG Tong, ZHANG Xiaonan, YU Xin, LIU Xiaofeng, and ZHAO Xiaochun, . Effect of R2R3-MYB Transcription Factor CitMYB21 on Flavonoids Biosynthesis in Citrus [J]. Acta Horticulturae Sinica, 2023, 50(2): 250-264. |
| [2] | GE Shibei, ZHANG Xuening, HAN Wenyan, LI Qingyun, LI Xin. Research Progress on Plant Flavonoids Biosynthesis and Their Anti-stress Mechanism [J]. Acta Horticulturae Sinica, 2023, 50(1): 209-224. |
| [3] | ZOU Xue, DING Fan, LIU Lifang, YU Hankaizong, CHEN Nianwei, and RAO Liping. A New Purple Potato Cultivar‘Mianziyu 1’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 93-94. |
| [4] | WANG Sha, ZHANG Xinhui, ZHAO Yujie, LI Bianbian, ZHAO Xueqing, SHEN Yu, DONG Jianmei, YUAN Zhaohe. Cloning and Functional Analysis of PgMYB111 Related to Anthocyanin Synthesis in Pomegranate [J]. Acta Horticulturae Sinica, 2022, 49(9): 1883-1894. |
| [5] | HUANG Ling, HU Xianmei, LIANG Zehui, WANG Yanping, CHAN Zhulong, XIANG Lin. Cloning and Function Identification of Anthocyanidin Synthase Gene TgANS in Tulipa gesneriana [J]. Acta Horticulturae Sinica, 2022, 49(9): 1935-1944. |
| [6] | LI Maofu, YANG Yuan, WANG Hua, FAN Youwei, SUN Pei, JIN Wanmei. Analysis the Function of R2R3 MYB Transcription Factor RhMYB113c on Regulating Anthocyanin Synthesis in Rosa hybrida [J]. Acta Horticulturae Sinica, 2022, 49(9): 1957-1966. |
| [7] | XU Haifeng, WANG Zhongtang, CHEN Xin, LIU Zhiguo, WANG Lihu, LIU Ping, LIU Mengjun, ZHANG Qiong. The Analyses of Target Metabolomics in Flavonoid and Its Potential MYB Regulation Factors During Coloring Period of Winter Jujube [J]. Acta Horticulturae Sinica, 2022, 49(8): 1761-1771. |
| [8] | QIAN Jieyu, JIANG Lingli, ZHENG Gang, CHEN Jiahong, LAI Wuhao, XU Menghan, FU Jianxin, ZHANG Chao. Identification and Expression Analysis of MYB Transcription Factors Regulating the Anthocyanin Biosynthesis in Zinnia elegans and Function Research of ZeMYB9 [J]. Acta Horticulturae Sinica, 2022, 49(7): 1505-1518. |
| [9] | ZHANG Lugang, LU Qianqian, HE Qiong, XUE Yihua, MA Xiaomin, MA Shuai, NIE Shanshan, YANG Wenjing. Creation of Novel Germplasm of Purple-orange Heading Chinese Cabbage [J]. Acta Horticulturae Sinica, 2022, 49(7): 1582-1588. |
| [10] | CHEN Daozong, LIU Yi, SHEN Wenjie, ZHU Bo, TAN Chen. Identification and Analysis of PAP1/2 Homologous Genes in Brassica rapa,B. oleracea and B. napus [J]. Acta Horticulturae Sinica, 2022, 49(6): 1301-1312. |
| [11] | LI Xiaoming, YU Junchi, WANG Chunxia. Comparison of Growth and Secondary Metabolites of Purple and White Flower Dracocephalum moldavica Under Field,Greenhouse and Greenhouse Shading Conditions [J]. Acta Horticulturae Sinica, 2022, 49(6): 1363-1370. |
| [12] | LI Lixian, WANG Shuo, CHEN Ying, WU Yingtao, WANG Yaqian, FANG Yue, CHEN Xuesen, TIAN Changping, FENG Shouqian. PavMYB10.1 Promotes Anthocyanin Accumulation by Positively Regulating PavRiant in Sweet Cherry [J]. Acta Horticulturae Sinica, 2022, 49(5): 1023-1030. |
| [13] | DENG Jiao, SU Mengyue, LIU Xuelian, OU Kefang, HU Zhengrong, YANG Pingfang. Transcriptome Analysis Revealed the Formation Mechanism of Floral Color of Lotus‘Dasajin’with Bicolor Petal [J]. Acta Horticulturae Sinica, 2022, 49(2): 365-377. |
| [14] | WANG Zhiyu, CHANG Beibei, LIU Qi, CHENG Xiaofan, DU Xiaoyun, YU Xiaoli, SONG Laiqing, ZHAO Lingling. Study on Expression and Anthocyanin Accumulation of Solute Carrier Gene MdSLC35F2-like in Apple [J]. Acta Horticulturae Sinica, 2022, 49(11): 2293-2303. |
| [15] | SU Yanli, GAO Xiaoming, YANG Jian, WANG Long, WANG Suke, ZHANG Xiangzhan, XUE Huabai. Dynamic Changes of Browning Degree,Phenolics Contents and Related Enzyme Activities During Pear Fruit Development [J]. Acta Horticulturae Sinica, 2022, 49(11): 2304-2312. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd