
Acta Horticulturae Sinica ›› 2021, Vol. 48 ›› Issue (7): 1317-1328.doi: 10.16420/j.issn.0513-353x.2020-0656
• Research Papars • Previous Articles Next Articles
YANG Shuangjuan, Zhang Xiaowei, WEI Xiaochun, ZHAO Yanyan, WANG Zhiyong, ZHAO Xiaobin, LI Lin, YUAN Yuxiang**( )
)
Received:2020-12-24
Revised:2021-02-19
Online:2021-07-25
Published:2021-08-10
Contact:
YUAN Yuxiang
E-mail:yuanyuxiang@126.com
CLC Number:
YANG Shuangjuan, Zhang Xiaowei, WEI Xiaochun, ZHAO Yanyan, WANG Zhiyong, ZHAO Xiaobin, LI Lin, YUAN Yuxiang. Mapping and KASP Markers Development for Clubroot Resistance Gene BraA.Pb.8.4 in Chinese Cabbage[J]. Acta Horticulturae Sinica, 2021, 48(7): 1317-1328.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2020-0656
| 样品名称 Sample | 高质量读长数 Clean reads | 高质量碱基数 Clean bases | 比对的读长数 Mapped reads | 比对率/% Mapping rate | 基因组覆盖倍数 Coverage of B. rapa genome |
|---|---|---|---|---|---|
| 1E519BC1F2-Rp | 146 359 698 | 21 953 954 700 | 139 582 489 | 95.37 | 45.27 |
| 1E519BC1F2-Sp | 130 400 096 | 19 560 014 400 | 124 262 099 | 95.29 | 40.33 |
| 4E511BC1F2-Rp | 171 165 594 | 25 674 839 100 | 163 000 999 | 95.23 | 52.94 |
| 4E511BC1F2-Sp | 125 843 472 | 18 876 520 800 | 119 448 979 | 94.92 | 38.92 |
Table 1 Summary of whole-genome re-sequencing data
| 样品名称 Sample | 高质量读长数 Clean reads | 高质量碱基数 Clean bases | 比对的读长数 Mapped reads | 比对率/% Mapping rate | 基因组覆盖倍数 Coverage of B. rapa genome |
|---|---|---|---|---|---|
| 1E519BC1F2-Rp | 146 359 698 | 21 953 954 700 | 139 582 489 | 95.37 | 45.27 |
| 1E519BC1F2-Sp | 130 400 096 | 19 560 014 400 | 124 262 099 | 95.29 | 40.33 |
| 4E511BC1F2-Rp | 171 165 594 | 25 674 839 100 | 163 000 999 | 95.23 | 52.94 |
| 4E511BC1F2-Sp | 125 843 472 | 18 876 520 800 | 119 448 979 | 94.92 | 38.92 |
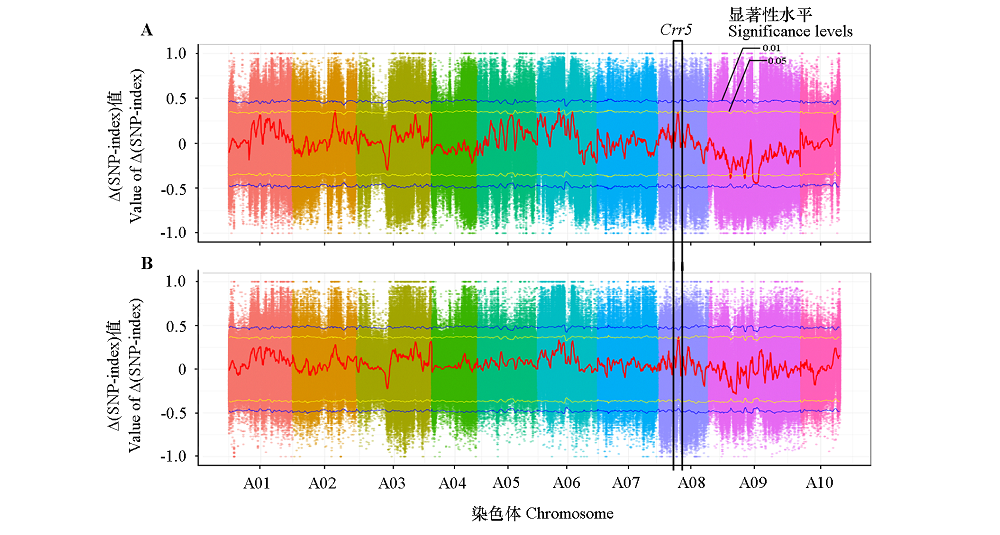
Fig. 3 BSA-seq analysis of 1E519BC1F2(A)and 4E511BC1F2(B)populations in Chinese cabbage The common candidate region in the two populations is named as BraA.Pb.8.4;the yellow and blue line in Manhattan plots indicate 0.05 and 0.01 significance levels,respectively.
| 群体 Population | 位置/Mb Location | 抗病混池SNP index Rp_SNP_index | 感病混池SNP index Sp_SNP_index | ∆SNP-index | P值 P-value |
|---|---|---|---|---|---|
| 1E519BC1F2 | A08:7.55 ~ 8.75 | 0.48 | 0.14 | 0.34 | < 0.05 |
| 4E511BC1F2 | A08:7.40 ~ 8.85 | 0.45 | 0.10 | 0.35 | < 0.05 |
Table 2 Information of the common candidate region in two populations
| 群体 Population | 位置/Mb Location | 抗病混池SNP index Rp_SNP_index | 感病混池SNP index Sp_SNP_index | ∆SNP-index | P值 P-value |
|---|---|---|---|---|---|
| 1E519BC1F2 | A08:7.55 ~ 8.75 | 0.48 | 0.14 | 0.34 | < 0.05 |
| 4E511BC1F2 | A08:7.40 ~ 8.85 | 0.45 | 0.10 | 0.35 | < 0.05 |
| KASP标记 KASP marker | 物理位置 Physical position | 变异位点 Variation locus | 引物序列(5′-3′) Primer sequence |
|---|---|---|---|
| BraA.Pb.8.4-K1 | A08:7 953 439 | T/C | Fa:GAAGGTGACCAAGTTCATGCTCGTGGTGGAAAGAAATGGTT |
| Fb:GAAGGTCGGAGTCAACGGATTCGTGGTGGAAAGAAATGGTC | |||
| R:CGTCCTCCGAGAAGCACACACC | |||
| BraA.Pb.8.4-K2 | A08:7 961 681 | T/G | Fa:GAAGGTCGGAGTCAACGGATTCGTCCTGCATTTCTTAGATGGT |
| Fb:GAAGGTGACCAAGTTCATGCTCGTCCTGCATTTCTTAGATGGG | |||
| R:CGAGCACTGATCTGCAAACACA | |||
| BraA.Pb.8.4-K3 | A08:7 970 088 | C/T | Fa:GAAGGTGACCAAGTTCATGCTGTTTAAGTAGAGACTGTGGGAC |
| Fb:GAAGGTCGGAGTCAACGGATTGTTTAAGTAGAGACTGTGGGAT | |||
| R:CTTCAAACGCAGCTTTCTTTGACTT |
Table 3 Information of KASP markers linked to gene BraA.Pb.8.4
| KASP标记 KASP marker | 物理位置 Physical position | 变异位点 Variation locus | 引物序列(5′-3′) Primer sequence |
|---|---|---|---|
| BraA.Pb.8.4-K1 | A08:7 953 439 | T/C | Fa:GAAGGTGACCAAGTTCATGCTCGTGGTGGAAAGAAATGGTT |
| Fb:GAAGGTCGGAGTCAACGGATTCGTGGTGGAAAGAAATGGTC | |||
| R:CGTCCTCCGAGAAGCACACACC | |||
| BraA.Pb.8.4-K2 | A08:7 961 681 | T/G | Fa:GAAGGTCGGAGTCAACGGATTCGTCCTGCATTTCTTAGATGGT |
| Fb:GAAGGTGACCAAGTTCATGCTCGTCCTGCATTTCTTAGATGGG | |||
| R:CGAGCACTGATCTGCAAACACA | |||
| BraA.Pb.8.4-K3 | A08:7 970 088 | C/T | Fa:GAAGGTGACCAAGTTCATGCTGTTTAAGTAGAGACTGTGGGAC |
| Fb:GAAGGTCGGAGTCAACGGATTGTTTAAGTAGAGACTGTGGGAT | |||
| R:CTTCAAACGCAGCTTTCTTTGACTT |

Fig. 4 KASP genotyping of 1E519BC1F2 population with primers BraA.Pb.8.4-K1(A,B)and BraA.Pb.8.4-K2(C,D) A,C:KASP genotyping result in 96-well plate format;B,D:KASP genotyping result in X-Y coordinates.
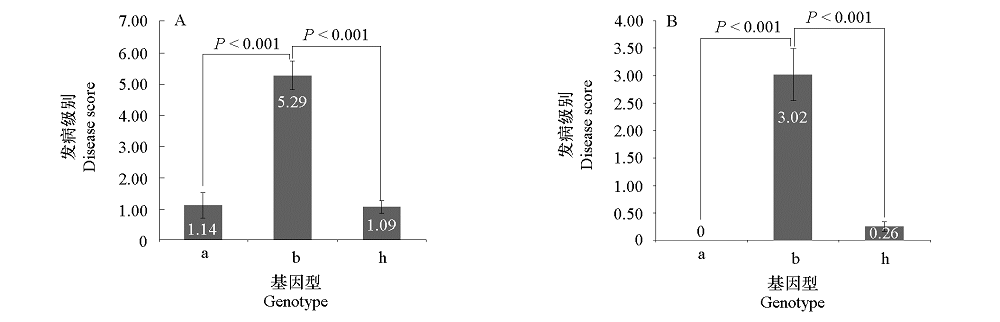
Fig. 5 Correlation analysis between genotypes of marker BraA.Pb.8.4-K1 and clubroot resistance phenotypes in population 1E519BC1F2(A)and 4E511BC1F2(B)
| 群体 Population | 根肿病抗性 Clubroot resistance | 单株数 Number of individual | 基因型 Genotype | 一致率/% Concordance rate | ||
|---|---|---|---|---|---|---|
| a | b | h | ||||
| 1E177BC2F2-5 | 抗病Resistant | 19 | 0 | 1 | 18 | 94.74 |
| 感病Susceptible | 19 | 0 | 18 | 1 | ||
| 1E177BC2F2-24 | 抗病Resistant | 28 | 9 | 0 | 19 | 100.00 |
| 感病Susceptible | 11 | 0 | 11 | 0 | ||
| 1E177BC2F2-25 | 抗病Resistant | 34 | 9 | 2 | 23 | 91.30 |
| 感病Susceptible | 12 | 0 | 10 | 2 | ||
| 1E177BC2F2-26 | 抗病Resistant | 29 | 6 | 0 | 23 | 100.00 |
| 感病Susceptible | 12 | 0 | 12 | 0 | ||
| CR1E-10 | 抗病Resistant | 45 | 0 | 5 | 40 | 94.62 |
| 感病Susceptible | 48 | 0 | 48 | 0 | ||
Table 4 Validation of marker BraA.Pb.8.4-K1 in five populations
| 群体 Population | 根肿病抗性 Clubroot resistance | 单株数 Number of individual | 基因型 Genotype | 一致率/% Concordance rate | ||
|---|---|---|---|---|---|---|
| a | b | h | ||||
| 1E177BC2F2-5 | 抗病Resistant | 19 | 0 | 1 | 18 | 94.74 |
| 感病Susceptible | 19 | 0 | 18 | 1 | ||
| 1E177BC2F2-24 | 抗病Resistant | 28 | 9 | 0 | 19 | 100.00 |
| 感病Susceptible | 11 | 0 | 11 | 0 | ||
| 1E177BC2F2-25 | 抗病Resistant | 34 | 9 | 2 | 23 | 91.30 |
| 感病Susceptible | 12 | 0 | 10 | 2 | ||
| 1E177BC2F2-26 | 抗病Resistant | 29 | 6 | 0 | 23 | 100.00 |
| 感病Susceptible | 12 | 0 | 12 | 0 | ||
| CR1E-10 | 抗病Resistant | 45 | 0 | 5 | 40 | 94.62 |
| 感病Susceptible | 48 | 0 | 48 | 0 | ||
| 基因 Gene | 标记或候选基因 Marker or candidate gene | A08物理位置/bp Physical position on A08 |
|---|---|---|
| BraA.Pb.8.4 | BraA.Pb.8.4-K1 | 7 953 439 |
| BraA.Pb.8.4-K2 | 7 961 681 | |
| BraA.Pb.8.4-K3 | 7 970 088 | |
| CRs | Probe55 | 9 956 858 |
| Probe59 | 10 497 705 | |
| Probe60 | 10 539 495 | |
| Probe64 | 10 707 395 | |
| Rcr9 | Bra020936 | 10 295 068 ~ 10 295 637 |
| Crr1 | Bra020861 | 10 809 433 ~ 10 825 238 |
Table 5 Comparison the physical position of BraA.Pb.8.4 and other genes in chromosome A08
| 基因 Gene | 标记或候选基因 Marker or candidate gene | A08物理位置/bp Physical position on A08 |
|---|---|---|
| BraA.Pb.8.4 | BraA.Pb.8.4-K1 | 7 953 439 |
| BraA.Pb.8.4-K2 | 7 961 681 | |
| BraA.Pb.8.4-K3 | 7 970 088 | |
| CRs | Probe55 | 9 956 858 |
| Probe59 | 10 497 705 | |
| Probe60 | 10 539 495 | |
| Probe64 | 10 707 395 | |
| Rcr9 | Bra020936 | 10 295 068 ~ 10 295 637 |
| Crr1 | Bra020861 | 10 809 433 ~ 10 825 238 |
| 标记 Marker | 正向引物序列(5′-3′) Forward primer | 反向引物序列(5′-3′) Reverse primer | 文献来源 Reference |
|---|---|---|---|
| Probe55 | ATTCAGATGACCTTCTTCCGAC | GATATGTTACCACGGCGAAAAC | Laila et al.,2019 |
| Probe59 | ACAGCAAAACCTCTCTTCTCTC | ATGTCAAACAAGCGTCTAAAGC | Laila et al.,2019 |
| Probe60 | ACACTTAGGTGAAGAAGCAACA | GCATGCATCAAGTAACTTGCAA | Laila et al.,2019 |
| Probe 64 | GTTGGAGAACACACTGAGATCA | TCTCTCGAGGCTTCTGTTACTA | Laila et al.,2019 |
| Rcr9_specific | AGTGGAGAACCAAAGCCAAA | CAATTGTGGCCACCTTCTTC | Yu et al.,2017 |
| BAS2 | TGTTAGACATCAAAGAGGGCTTGAG | TCTCAAAGACGATAATGAACCCAAA | Suwabe et al.,2012 |
| BAS7 | AAGCAGAAGGGTTCTTGTCTGAACT | TCACTCTGAGACATGAGGACACTTG | Suwabe et al.,2012 |
Table 6 Primers used for validation the novelty of BraA.Pb.8.4
| 标记 Marker | 正向引物序列(5′-3′) Forward primer | 反向引物序列(5′-3′) Reverse primer | 文献来源 Reference |
|---|---|---|---|
| Probe55 | ATTCAGATGACCTTCTTCCGAC | GATATGTTACCACGGCGAAAAC | Laila et al.,2019 |
| Probe59 | ACAGCAAAACCTCTCTTCTCTC | ATGTCAAACAAGCGTCTAAAGC | Laila et al.,2019 |
| Probe60 | ACACTTAGGTGAAGAAGCAACA | GCATGCATCAAGTAACTTGCAA | Laila et al.,2019 |
| Probe 64 | GTTGGAGAACACACTGAGATCA | TCTCTCGAGGCTTCTGTTACTA | Laila et al.,2019 |
| Rcr9_specific | AGTGGAGAACCAAAGCCAAA | CAATTGTGGCCACCTTCTTC | Yu et al.,2017 |
| BAS2 | TGTTAGACATCAAAGAGGGCTTGAG | TCTCAAAGACGATAATGAACCCAAA | Suwabe et al.,2012 |
| BAS7 | AAGCAGAAGGGTTCTTGTCTGAACT | TCACTCTGAGACATGAGGACACTTG | Suwabe et al.,2012 |
| [1] | Chen J, Jing J, Zhan Z, Zhang T, Zhang C, Piao Z. 2013. Identification of novel QTLs for isolate-specific partial resistance to Plasmodiophora brassicae in Brassica rapa. PLoS ONE, 8:e85307. |
| [2] |
Chu M, Song T, Falk K C, Zhang X, Liu X, Chang A, Lahlali R, McGregor L, Gossen B D, Yu F. 2014. Fine mapping of Rcr1 and analyses of its effect on transcriptome patterns during infection by Plasmodiophora brassicae. BMC Genomics, 15:1166.
doi: 10.1186/1471-2164-15-1166 URL |
| [3] |
Hatakeyama K, Suwabe K, Tomita R N, Kato T, Nunome T, Fukuoka H, Matsumoto S. 2013. Identification and characterization of Crr1a,a gene for resistance to clubroot disease(Plasmodiophora brassicae Woronin.)in Brassica rapa L. PLoS ONE, 8:e54745.
doi: 10.1371/journal.pone.0054745 URL |
| [4] |
Hirai M, Harada T, Kubo N, Tsukada M, Suwabe K, Matsumoto S. 2004. A novel locus for clubroot resistance in Brassica rapa and its linkage markers. Theoretical Applied Genetics, 108:639-643.
doi: 10.1007/s00122-003-1475-x URL |
| [5] |
Laila R, Park J I, Robin A H K, Natarajan S, Vijayakumar H, Shirasawa K, Isobe S, Kim H T, Nou I S. 2019. Mapping of a novel clubroot resistance QTL using ddRAD-seq in Chinese cabbage(Brassica rapa L.). BMC Plant Biology, 19(1):13.
doi: 10.1186/s12870-018-1615-8 URL |
| [6] |
Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25:1754-1760.
doi: 10.1093/bioinformatics/btp324 URL |
| [7] |
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics, 25:2078-2079.
doi: 10.1093/bioinformatics/btp352 URL |
| [8] | Long Tong, Zhang Yani, Yang Nina, Cao Chunxia, Zhang Guangyang, Zhang Shilong, Cheng Xianliang, Huang Daye, Wan Zhongyi. 2019. Progress in biological control of Chinese cabbage clubroot disease. Hubei Agricultural Sciences, 58(5):5-8. (in Chinese) |
| 龙同, 张亚妮, 杨妮娜, 曹春霞, 张光阳, 张士龙, 程贤亮, 黄大野, 万中义. 2019. 大白菜根肿病生物防治研究进展. 湖北农业科学, 58(5):5-8. | |
| [9] |
Matsumoto E, Yasui C, Ohi M, Tsukada M. 1998. Linkage analysis of RFLP markers for clubroot resistance and pigmentation in Chinese cabbage(Brassica rapa ssp. pekinensis). Euphytica, 104:79.
doi: 10.1023/A:1018370418201 URL |
| [10] |
Pang W, Fu P, Li X, Zhan Z, Yu S, Piao Z. 2018. Identification and mapping of the clubroot resistance gene CRd in Chinese cabbage(Brassica rapa L. ssp. pekinensis). Frontiers in Plant Science, 9:653.
doi: 10.3389/fpls.2018.00653 URL |
| [11] |
Piao Z, Deng Y, Choi S, Park Y, Lim Y. 2004. SCAR and CAPS mapping of CRb,a gene conferring resistance to Plasmodiophora brassicae in Chinese cabbage(Brassica rapa ssp. pekinensis). Theoretical Applied Genetics, 108:1458-1465.
doi: 10.1007/s00122-003-1577-5 URL |
| [12] |
Sakamoto K, Saito A, Hayashida N, Taguchi G, Matsumoto E. 2008. Mapping of isolate-specific QTLs for clubroot resistance in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Theoretical Applied Genetics, 117:759-767.
doi: 10.1007/s00122-008-0817-0 URL |
| [13] |
Semagn K, Babu R, Hearne S, Olsen M. 2014. Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR(KASP):Overview of the technology and its application in crop improvement. Molecular Breeding, 33(1):1-14.
doi: 10.1007/s11032-013-9917-x URL |
| [14] | Sun Baoya, Shen Xiangqun, Guo Haifeng, Zhou Yonghong. 2005. Research progress in clubroot of crucifers and resistance breeding. China Vegetables, (4):34-37. (in Chinese) |
| 孙保亚, 沈向群, 郭海峰, 周永红. 2005. 十字花科植物根肿病及抗病育种研究进展. 中国蔬菜, (4):34-37. | |
| [15] |
Suwabe K, Suzuki G, Nunome T, Hatakeyama K, Mukai Y, Fukuoka H, Matsumoto S. 2012. Microstructure of a Brassica rapa genome segment homoeologous to the resistance gene cluster on Arabidopsis chromosome 4. Breeding Science, 62:170-177.
doi: 10.1270/jsbbs.62.170 URL pmid: 23136528 |
| [16] |
Suwabe K, Tsukazaki H, Iketani H, Hatakeyama K, Fujimura M, Nunome T, Fukuoka H, Matsumoto S, Hirai M. 2003. Identification of two loci for resistance to clubroot(Plasmodiophora brassicae Woron.)in Brassica rapa L. Theoretical Applied Genetics, 107:997-1002.
doi: 10.1007/s00122-003-1309-x URL |
| [17] |
Suwabe K, Tsukazaki H, Iketani H, Hatakeyama K, Kondo M, Fujimura M, Nunome T, Fukuoka H, Hirai M, Matsumoto S. 2006. Simple sequence repeat-based comparative genomics between Brassica rapa and Arabidopsis thaliana:the genetic origin of clubroot resistance. Genetics, 173:309-319.
pmid: 16723420 |
| [18] |
Takagi H, Abe A, Yoshida K, Kosugi S, Natsume S. 2013. QTLseq:rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. The Plant Journal, 74:174-183.
doi: 10.1111/tpj.2013.74.issue-1 URL |
| [19] |
Wang X Brassica rapa Genome Sequencing Project Consortium. 2011. The genome of the mesopolyploid crop species Brassica rapa. Nature Genetics, 43:1035-1039.
doi: 10.1038/ng.919 URL |
| [20] |
Wei X C, Xu W, Yuan Y X, Yao Q J, Zhao Y Y, Wang Z Y, Jiang W S, Zhang X W. 2016. Genome-wide investigation of microRNAs and their targets in Brassica rapa ssp. pekinensis root with Plasmodiophora brassicae infection. Horticultural Plant Journal, 2(4):209-216.
doi: 10.1016/j.hpj.2016.11.004 URL |
| [21] | Yang Limei, Fang Zhiyuan, Zhang Yangyong, Zhuang Mu, Lü Honghao, Wang Yong, Ji Jialei, Liu Yumei, Li Zhansheng, Han Fengqing. 2020. Recent advances of disease and stress resistance breeding of cabbage in China. Acta Horticulturae Sinica, 47(9):1678-1688. (in Chinese) |
| 杨丽梅, 方智远, 张扬勇, 庄木, 吕红豪, 王勇, 季家磊, 刘玉梅, 李占省, 韩风庆. 2020. 中国结球甘蓝抗病抗逆遗传育种近年研究进展. 园艺学报, 47(9):1678-1688. | |
| [22] | Yang Shaungjuan, Yuan Yuxiang, Wei Xiaochun, Wang Zhiyong, Zhao Yanyan, Yao Qiuju, Zhang Xiaowei. 2018. Optimization and establishment of KASP reaction system for Chinese cabbage(Brassica rapa L. ssp. pekinensis). Acta Horticulturae Sinica, 45(12):2442-2452. (in Chinese) |
| 杨双娟, 原玉香, 魏小春, 王志勇, 赵艳艳, 姚秋菊, 张晓伟. 2018. 大白菜 KASP 反应体系的优化与建立. 园艺学报, 45(12):2442-2452. | |
| [23] |
Yu F, Zhang X, Peng G, Falk K C, Strelkov S E, Gossen B D. 2017. Genotyping by sequencing reveals three QTL for clubroot resistance to six pathotypes of Plasmodiophora brassicae in Brassica rapa. Scientific Reports, 7(1):4516.
doi: 10.1038/s41598-017-04903-2 URL |
| [24] | Yuan Yuxiang, Zhao Yanyan, Wei Xiaochun, Yao Qiuju, Jiang Wusheng, Wang Zhiyong, Li Yang, Xu Qian, Yang Shuangjuan, Zhang Xiaowei. 2017. Pathotype identification of Plasmodiophora brassicae Woron. collected from Chinese cabbage in Henan Province. Journal of Henan Agricultural Sciences, 46(7):71-76. (in Chinese) |
| 原玉香, 赵艳艳, 魏小春, 姚秋菊, 蒋武生, 王志勇, 李扬, 许茜, 杨双娟, 张晓伟. 2017. 河南省大白菜根肿病菌生理小种鉴定. 河南农业科学, 46(7):71-76. | |
| [25] | Zhang Hui, Zhang Shujiang, Li Fei, Zhang Shifan, Li Guoliang, Ma Xiaochao, Liu Xitong, Sun Rifei. 2020. Research progress on clubroot disease resistance breeding of Brassica rapa. Acta Horticulturae Sinica, 47(9):1648-1662. (in Chinese) |
| 张慧, 张淑江, 李菲, 章时蕃, 李国亮, 马小超, 刘希童, 孙日飞. 2020. 大白菜抗根肿病育种研究进展. 园艺学报, 47(9):1648-1662. | |
| [26] | Zhang Jing, Wu Yue, Feng Hui, Ge Wenjie, Liu Xuyao, Lü Mingcan, Wang Yilian, Ji Ruiqin. 2019. Obtaining and identification of single-spore physiological races of Plasmodiophora brassicae in Williams system. Acta Horticulturae Sinica, 46(12):2415-2422. (in Chinese) |
| 张晶, 武月, 冯辉, 葛文杰, 刘旭垚, 吕明灿, 王艺琏, 冀瑞琴. 2019. 根肿菌Williams系统中各单孢生理小种的获得与鉴定. 园艺学报, 46(12):2415-2422. |
| [1] | HAN Shuhui, HAN Caifeng, HAN Shurong, HAN Caimei, and HAN Xu. A New Autumn Chinese Cabbage Hybrid‘Jiaoyan Qiubao’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 83-84. |
| [2] | WANG Weihong, ZHANG Fenglan, YU Yangjun, ZHANG Deshuang, ZHAO Xiuyun, YU Shuancang, SU Tongbing, LI Peirong, and XIN Xiaoyun. A New Chinese Cabbage Cultivar‘Jingqiu 1518’for Autumn Cultivation [J]. Acta Horticulturae Sinica, 2022, 49(S2): 85-86. |
| [3] | YU Yangjun, WANG Weihong, SU Tongbing, ZHANG Fenglan, ZHANG Deshuang, ZHAO Xiuyun, YU Shuancang, LI Peirong, XIN Xiaoyun, and WANG Jiao. A New Chinese Cabbage Cultivar‘Jingchun CR3’with Clubroot Resistance and Bolting Tolerance [J]. Acta Horticulturae Sinica, 2022, 49(S2): 87-88. |
| [4] | WANG Lili, WANG Xin, WU Haidong, WEN Qiang, and Yang Xiaofei. A New Chinese Cabbage Cultivar‘Liaobai 28’with Resistance to Clubroot Disease [J]. Acta Horticulturae Sinica, 2022, 49(S2): 89-90. |
| [5] | YU Yangjun, SU Tongbing, ZHANG Fenglan, ZHANG Deshuang, ZHAO Xiuyun, YU Shuancang, WANG Weihong, LI Peirong, XIN Xiaoyun, WANG Jiao, and WU Changjian. A New Purple Seedling-Edible Chinese Cabbage F1 Hybrid‘Jingyan Zikuaicai’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 91-92. |
| [6] | HUANG Li, CHEN Caizhi, YU Xiaolin, YAO Xiangtan, and CAO Jiashu, . A New Early-mid Maturing Chinese Cabbage Pak-choi Cultivar‘Zhedaqing’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 93-94. |
| [7] | XU Ligong, HAN Taili, SUN Jifeng, YANG Xiaodong, and TAN Jinxia. A New Seedling-edible Chinese Cabbage Cultivar‘Jinlü 2’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 67-68. |
| [8] | WANG Yu, ZHANG Xue, ZHANG Xueying, ZHANG Siyu, WEN Tingting, WANG Yingjun, GAN Caixia, PANG Wenxing. The Effect of Camalexin on Chinese Cabbage Resistance to Clubroot [J]. Acta Horticulturae Sinica, 2022, 49(8): 1689-1698. |
| [9] | ZHANG Lugang, LU Qianqian, HE Qiong, XUE Yihua, MA Xiaomin, MA Shuai, NIE Shanshan, YANG Wenjing. Creation of Novel Germplasm of Purple-orange Heading Chinese Cabbage [J]. Acta Horticulturae Sinica, 2022, 49(7): 1582-1588. |
| [10] | RUI Tingting, GAO Qingyun, LI Xiaojing, SHI Yanxia, XIE Xuewen, LI Lei, ZHANG Hongjie, XU Wenjuan, CHAI Ali, LI Baoju. Methylene Blue Combined Agarose Assay for Single-spore Isolation of Plasmodiophora brassicae and Pathotype Differentiation [J]. Acta Horticulturae Sinica, 2022, 49(6): 1290-1300. |
| [11] | QIAO Jun, LIU Jing, LI Suwen, WANG Liying. Prediction of Fruit Color Genes Under the Calyx of Eggplant Based on Genome-wide Resequencing in an Extreme Mixing Pool [J]. Acta Horticulturae Sinica, 2022, 49(3): 613-621. |
| [12] | WANG Guangpeng, LIU Tongkun, XU Xinfeng, LI Zhubo, GAO Zhanyuan, HOU Xilin. Identification of LEA Family and Expression Analysis of Some Members Under Low-temperature Stress in Chinese Cabbage [J]. Acta Horticulturae Sinica, 2022, 49(2): 304-318. |
| [13] | SHENG Yunyan, YANG Limin, DAI Dongyang, ZHANG Jiaxin, WANG Ling, WANG Di, CAI Yi, TIAN Limei. Cytological Observation of Fruit Peduncle Abscission Zone and Preliminary Mapping of Mature Fruit Abscission AL3 gene in Melon [J]. Acta Horticulturae Sinica, 2022, 49(2): 341-351. |
| [14] | WANG Ronghua, WANG Shubin, LIU Shuantao, LI Qiaoyun, ZHANG Zhigang, WANG Lihua, ZHAO Zhizhong. Transcriptome Analysis of Waxy Near-isogenic Lines in Chinese Cabbage Floral Axis [J]. Acta Horticulturae Sinica, 2022, 49(1): 62-72. |
| [15] | XU Ligong, HAN Taili, SUN Jifeng, YANG Xiaodong, TAN Jinxia, SONG Yinhang, LI Meng, ZHOU Luhong, and SUN Shasha. A New Spring Chinese Cabbage Cultivar‘Weichun 22’ [J]. Acta Horticulturae Sinica, 2021, 48(S2): 2825-2826. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd