
Acta Horticulturae Sinica ›› 2021, Vol. 48 ›› Issue (5): 947-959.doi: 10.16420/j.issn.0513-353x.2020-0800
• Research Papers • Previous Articles Next Articles
NIU Xiqaing1, LUO Xiaoyun1, KANG Kaicheng2, HUANG Xianzhong1,2,*( ), HU Nengbing1, SUI Yihu1, AI Hao1
), HU Nengbing1, SUI Yihu1, AI Hao1
Received:2020-12-07
Revised:2021-03-21
Online:2021-05-25
Published:2021-06-07
Contact:
HUANG Xianzhong
E-mail:huangxz@ahstu.edu.cn
CLC Number:
NIU Xiqaing, LUO Xiaoyun, KANG Kaicheng, HUANG Xianzhong, HU Nengbing, SUI Yihu, AI Hao. Genome-wide Identification,Comparative Evolution and Expression Analysis of PEBP Gene Family from Capsicum annuum[J]. Acta Horticulturae Sinica, 2021, 48(5): 947-959.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2020-0800
| 基因 Gene | 上游引物序列(5′-3′) Forward primer sequence | 下游引物序列(5′-3′) Reverse primer sequence |
|---|---|---|
| CaMFT | GCTCCAAGTCCAAGTGAACC | CCGAGATCAAGATGCTGTGC |
| CaFT1 | CAATGCGCCAGACATAATTG | AGACGACGACCACCAGTACC |
| CaFT2 | GGTTCATATTGGAGGCAACG | TGCTGGGATATCTGTGACCA |
| CaFT3 | TGGTACTGATTTGAGGCCTTCT | CTCAAATTTGGGTTGCTAGGAC |
| CaFT4 | TGGTACAAATTTGAGGCCTTCT | GCATTTCCAAAGGTTACTCCTG |
| CaTFL1-1 | ACCTCCACTGGATTGTGACAG | CATTCTCTGCAGCAAACCTTC |
| CaTFL1-2 | TAGAGGGGCTTGTGAACCAC | TCTTCACCCCCAATCTCAAC |
| CaTFL1-3 | ACGGACATTCCAGGCACTAC | CAGAATCGATCCCTGGAGAG |
| CaTFL1-4 | AGATCCTGATGTTCCTGGCC | CTGAACTCCGACGTTTCTGC |
| CaUBI3 | TGTCCATCTGCTCTCTGTTG | CACCCCAAGCACAATAAGAC |
Table 1 qRT-PCR primers for expression analysis of PEBP in Capsicum annuum
| 基因 Gene | 上游引物序列(5′-3′) Forward primer sequence | 下游引物序列(5′-3′) Reverse primer sequence |
|---|---|---|
| CaMFT | GCTCCAAGTCCAAGTGAACC | CCGAGATCAAGATGCTGTGC |
| CaFT1 | CAATGCGCCAGACATAATTG | AGACGACGACCACCAGTACC |
| CaFT2 | GGTTCATATTGGAGGCAACG | TGCTGGGATATCTGTGACCA |
| CaFT3 | TGGTACTGATTTGAGGCCTTCT | CTCAAATTTGGGTTGCTAGGAC |
| CaFT4 | TGGTACAAATTTGAGGCCTTCT | GCATTTCCAAAGGTTACTCCTG |
| CaTFL1-1 | ACCTCCACTGGATTGTGACAG | CATTCTCTGCAGCAAACCTTC |
| CaTFL1-2 | TAGAGGGGCTTGTGAACCAC | TCTTCACCCCCAATCTCAAC |
| CaTFL1-3 | ACGGACATTCCAGGCACTAC | CAGAATCGATCCCTGGAGAG |
| CaTFL1-4 | AGATCCTGATGTTCCTGGCC | CTGAACTCCGACGTTTCTGC |
| CaUBI3 | TGTCCATCTGCTCTCTGTTG | CACCCCAAGCACAATAAGAC |
| 基因 Gene | 基因登录号 GenBank ID | 染色体 Chr. No. | 开放阅读 框长度/bp ORF | 氨基酸数/aa Amino acid | 分子量/kD Molecular weight | 等电点 pI | 亚细胞定位 Subcellular location |
|---|---|---|---|---|---|---|---|
| CaMFT | PHT88651.1 | Chr03 | 522 | 173 | 18.98 | 9.20 | 细胞质和细胞核 Cytoplasmic and nuclear |
| CaFT1 | PHT82416.1 | Chr05 | 531 | 176 | 19.69 | 6.82 | 细胞质和细胞核 Cytoplasmic and nuclear |
| CaFT2 | PHT82541.1 | Chr05 | 528 | 175 | 19.81 | 7.76 | 细胞质和细胞核 Cytoplasmic and nuclear |
| CaFT3 | PHT65068.1 | Chr12 | 534 | 177 | 19.98 | 8.64 | 细胞质和细胞核 Cytoplasmic and nuclear |
| CaFT4 | PHT61065.1 | Sca. 2174 | 531 | 176 | 19.71 | 6.73 | 细胞质和细胞核 Cytoplasmic and nuclear |
| CaTFL1-1 | PHT94378.1 | Chr01 | 534 | 177 | 19.92 | 9.01 | 细胞质和细胞核 Cytoplasmic and nuclear |
| CaTFL1-2 | PHT87214.1 | Chr03 | 528 | 175 | 19.54 | 9.07 | 细胞质和细胞核 Cytoplasmic and nuclear |
| CaTFL1-3 | PHT80015.1 | Chr06 | 528 | 175 | 19.93 | 8.64 | 细胞质和细胞核 Cytoplasmic and nuclear |
| CaTFL1-4 | PHT71587.1 | Chr09 | 519 | 172 | 19.55 | 7.97 | 细胞质和细胞核 Cytoplasmic and nuclear |
Table 2 The information of PEBP gene family in Capsicum annuum
| 基因 Gene | 基因登录号 GenBank ID | 染色体 Chr. No. | 开放阅读 框长度/bp ORF | 氨基酸数/aa Amino acid | 分子量/kD Molecular weight | 等电点 pI | 亚细胞定位 Subcellular location |
|---|---|---|---|---|---|---|---|
| CaMFT | PHT88651.1 | Chr03 | 522 | 173 | 18.98 | 9.20 | 细胞质和细胞核 Cytoplasmic and nuclear |
| CaFT1 | PHT82416.1 | Chr05 | 531 | 176 | 19.69 | 6.82 | 细胞质和细胞核 Cytoplasmic and nuclear |
| CaFT2 | PHT82541.1 | Chr05 | 528 | 175 | 19.81 | 7.76 | 细胞质和细胞核 Cytoplasmic and nuclear |
| CaFT3 | PHT65068.1 | Chr12 | 534 | 177 | 19.98 | 8.64 | 细胞质和细胞核 Cytoplasmic and nuclear |
| CaFT4 | PHT61065.1 | Sca. 2174 | 531 | 176 | 19.71 | 6.73 | 细胞质和细胞核 Cytoplasmic and nuclear |
| CaTFL1-1 | PHT94378.1 | Chr01 | 534 | 177 | 19.92 | 9.01 | 细胞质和细胞核 Cytoplasmic and nuclear |
| CaTFL1-2 | PHT87214.1 | Chr03 | 528 | 175 | 19.54 | 9.07 | 细胞质和细胞核 Cytoplasmic and nuclear |
| CaTFL1-3 | PHT80015.1 | Chr06 | 528 | 175 | 19.93 | 8.64 | 细胞质和细胞核 Cytoplasmic and nuclear |
| CaTFL1-4 | PHT71587.1 | Chr09 | 519 | 172 | 19.55 | 7.97 | 细胞质和细胞核 Cytoplasmic and nuclear |
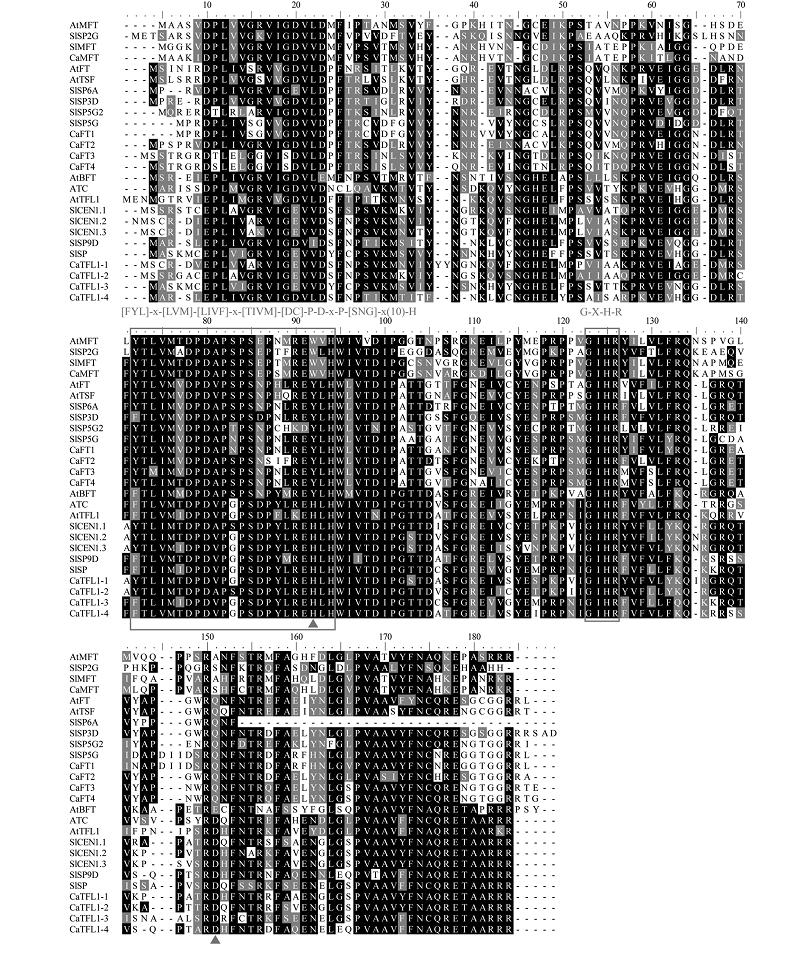
Fig. 1 Multiple amino acid sequence alignment of PEBP proteins from Arabidopsis thaliana,Solanum lycopersicum and Capsicum annuum The red triangles represent the key amino acids that determine the functions of FT-like and TFL1-like,the rectangles represent [FYL]-x-[LVM]-[LIVF]-X-[TIVM]-[DC]-P-d-x-P-[SNG]-x(10)-H and G-X-H-R conserved motif. At:Arabidopsis thaliana;Sl:Solanum lycopersicum;Ca:Capsicum annuum.
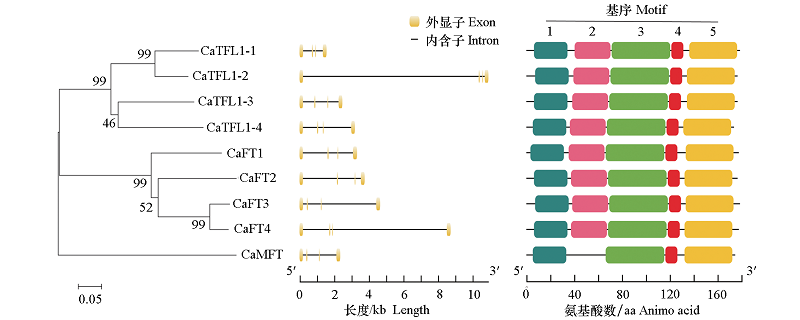
Fig. 2 Phylogentic tree of CaPEBP,gene intron/exon structure and conserved motif analysis of the PEBP genes in Capsicum annuum The number in the tree represents boots value. The scale bar represents branch length.
| 基序 Motif | 长度/aa Length | 氨基酸保守序列 Amino acid conserved sequence |
|---|---|---|
| motif 1 | 29 | DPLVVGRVIGDVVDPFTPSVKMSVVYNNR |
| motif 2 | 31 | YNGHELRPSQIAAQPRVEIGGNDLRTFYTLI |
| motif 3 | 50 | MTDPDAPSPSBPYLREYLHWJVTDIPGTTDVSFGNEIVCYESPRPSIGIH |
| motif 4 | 11 | RYVFVLFRQLG |
| motif 5 | 41 | VYAPTWRDHFNTRDFAEENNLGSPVAAVYFNCQRETATRRR |
Table 3 Conserved motifs of the PEBP proteins in Capsicum annuum
| 基序 Motif | 长度/aa Length | 氨基酸保守序列 Amino acid conserved sequence |
|---|---|---|
| motif 1 | 29 | DPLVVGRVIGDVVDPFTPSVKMSVVYNNR |
| motif 2 | 31 | YNGHELRPSQIAAQPRVEIGGNDLRTFYTLI |
| motif 3 | 50 | MTDPDAPSPSBPYLREYLHWJVTDIPGTTDVSFGNEIVCYESPRPSIGIH |
| motif 4 | 11 | RYVFVLFRQLG |
| motif 5 | 41 | VYAPTWRDHFNTRDFAEENNLGSPVAAVYFNCQRETATRRR |
| 辣椒基因 Gene ID in C. annuum | 番茄基因 Gene ID in S. lycopersicum | 基因模型 Gene model | 非同义突变频/Ka | 同义突变频率/Ks | Ka/Ks |
|---|---|---|---|---|---|
| CaMFT | SlMFT | MFT-like | 0.07 | 0.44 | 0.17 |
| CaFT1 | SlSP5G | FT-like | 0.03 | 0.39 | 0.09 |
| CaFT2 | SlSP6A | FT-like | 0.06 | 0.24 | 0.27 |
| CaFT3 | SlSP5G3 | FT-like | 0.03 | 0.29 | 0.11 |
| CaFT4 | SlSP5G1 | FT-like | 0.18 | 0.48 | 0.37 |
| CaTFL1-1 | SlCEN1.3 | TFL1-like | 0.06 | 0.56 | 0.11 |
| CaTFL1-2 | SlCEN1.1 | TFL1-like | 0.04 | 0.57 | 0.08 |
| CaTFL1-3 | SlSP | TFL1-like | 0.03 | 0.22 | 0.15 |
| SlCEN1.3 | TFL1-like | 0.26 | 1.16 | 0.22 | |
| CaTFL1-4 | SlSP9D | TFL1-like | 0.05 | 0.36 | 0.13 |
Table 4 Ka/Ks values of PEBP genes between Capsicum annuum and Solanum lycopersicum
| 辣椒基因 Gene ID in C. annuum | 番茄基因 Gene ID in S. lycopersicum | 基因模型 Gene model | 非同义突变频/Ka | 同义突变频率/Ks | Ka/Ks |
|---|---|---|---|---|---|
| CaMFT | SlMFT | MFT-like | 0.07 | 0.44 | 0.17 |
| CaFT1 | SlSP5G | FT-like | 0.03 | 0.39 | 0.09 |
| CaFT2 | SlSP6A | FT-like | 0.06 | 0.24 | 0.27 |
| CaFT3 | SlSP5G3 | FT-like | 0.03 | 0.29 | 0.11 |
| CaFT4 | SlSP5G1 | FT-like | 0.18 | 0.48 | 0.37 |
| CaTFL1-1 | SlCEN1.3 | TFL1-like | 0.06 | 0.56 | 0.11 |
| CaTFL1-2 | SlCEN1.1 | TFL1-like | 0.04 | 0.57 | 0.08 |
| CaTFL1-3 | SlSP | TFL1-like | 0.03 | 0.22 | 0.15 |
| SlCEN1.3 | TFL1-like | 0.26 | 1.16 | 0.22 | |
| CaTFL1-4 | SlSP9D | TFL1-like | 0.05 | 0.36 | 0.13 |
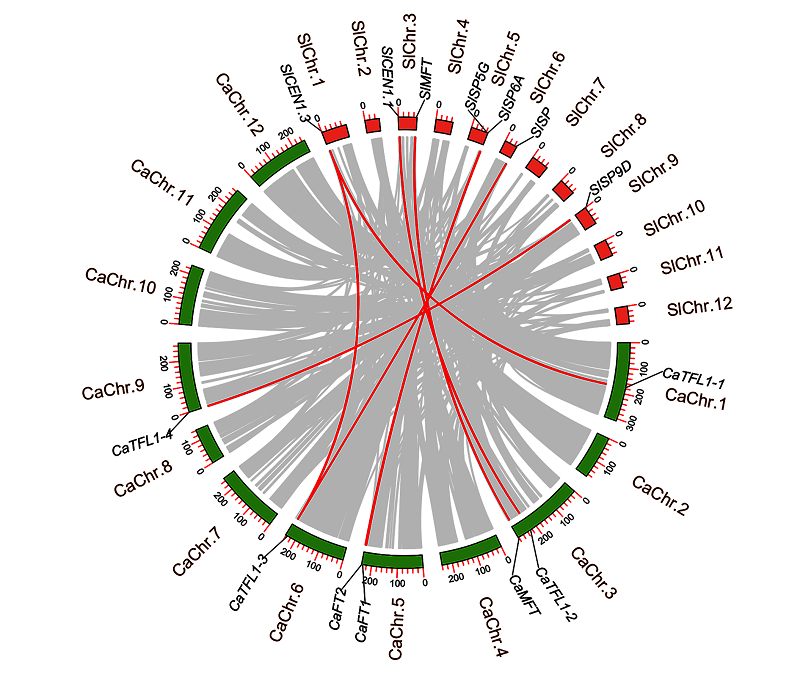
Fig. 3 Collinearity analysis of orthologous PEBP gene pairs betweenSolanum lycopersicum andCapsicum annuum The red lines representPEBP collinearity between species,and the gray lines are collinearity of all gene members between species.
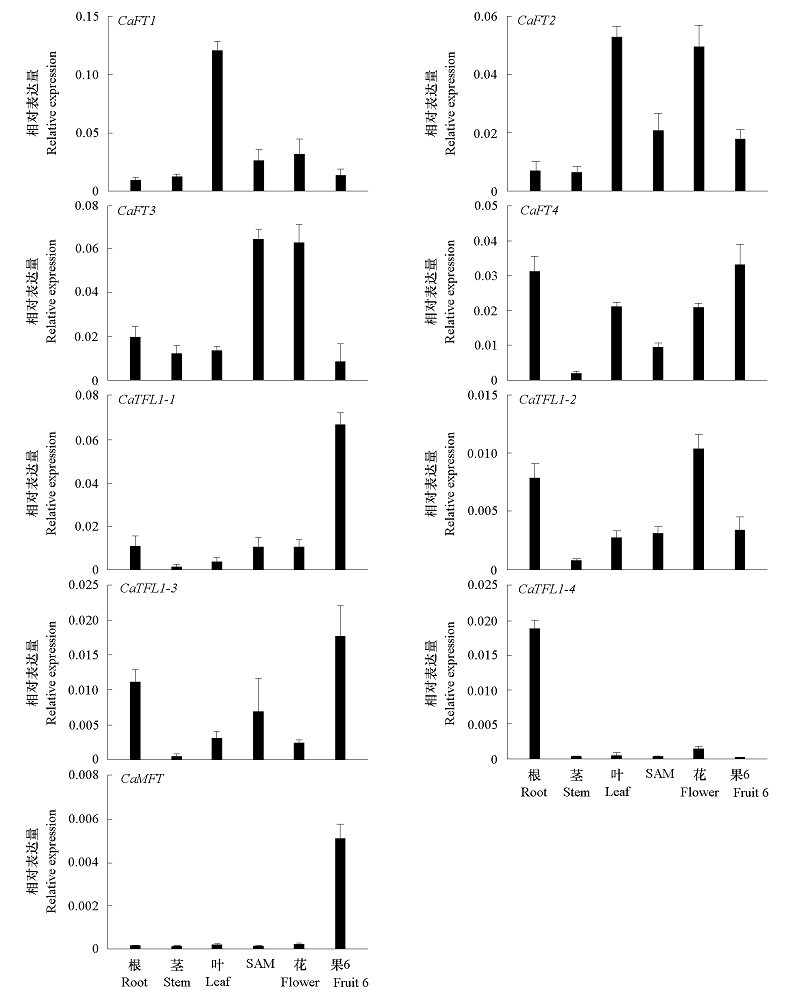
Fig. 4 Expression analysis of CaPEBP genes by qRT-PCR in roots,stems,leaves,shoot apical meristems(SAM),flowers and fruit(6th stage of fruit development)of pepper
| [1] |
Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. 2005. FD,a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science, 309(5737):1052-1056.
doi: 10.1126/science.1115983 URL |
| [2] |
Ahn J H, Miller D, Winter V J, Banfield M J, Lee J H, Yoo S Y, Henz S R, Brady R L, Weigel D. 2006. A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. The EMBO Journal, 25(3):605-614.
doi: 10.1038/sj.emboj.7600950 URL |
| [3] | Bernier I, Jolles P. 1984. Purification and characterisation of a basic 23 kDa cytosolic protein from bovine brain. Biochimica et Biophysica Acta, 790(2):174-181. |
| [4] | Cao K, Cui L, Zhou X, Ye L, Zou Z, Deng S. 2016. Four tomato FLOWERING LOCUS T-like proteins act antagonistically to regulate floral initiation. Frontiers in Plant Science, 6:1213. |
| [5] |
Carmel-Goren L, Liu Y S, Lifschitz E, Zamir D. 2003. The SELF-PRUNING gene family in tomato. Plant Molecular Biology, 52:1215-1222.
doi: 10.1023/B:PLAN.0000004333.96451.11 URL |
| [6] |
Carmona M J, Calonje M, Martínez-Zapater J M. 2007. The FT/TFL1 gene family in grapevine. Plant Molecular Biology, 63(5):637-650.
doi: 10.1007/s11103-006-9113-z URL |
| [7] |
Chardon F, Damerval C. 2005. Phylogenomic analysis of the PEBP gene family in cereals. Journal of Molecular Evolution, 61(5):579-590.
doi: 10.1007/s00239-004-0179-4 URL |
| [8] |
Chen C, Chen H, Zhang Y, Thomas H R, Frank M H, He Y, Xia R. 2020. TBtools:an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 13(8):1194-1202.
doi: 10.1016/j.molp.2020.06.009 URL |
| [9] |
Elitzur T, Nahum H, Borovsky Y, Pekker I, Eshed Y, Paran I. 2009. Co-ordinated regulation of flowering time,plant architecture and growth by FASCICULATE:the pepper orthologue of SELF PRUNING. Journal of Experimental Botany, 60(3):869-880.
doi: 10.1093/jxb/ern334 URL |
| [10] |
Eshed Y, Lippman Z B. 2019. Revolutions in agriculture chart a course for targeted breeding of old and new crops. Science, 366(6466):eaax0025.
doi: 10.1126/science.aax0025 URL |
| [11] | Hanzawa Y, Money T, Bradley D. 2005. A single amino acid converts a repressor to an activator of flowering. Proceedings of the National Academy of Science of the United States of America, 102(21):7748-7753. |
| [12] |
Hedman H, Källman T, Lagercrantz U. 2009. Early evolution of the MFT-like gene family in plants. Plant Molecular Biology, 70(4):359-369.
doi: 10.1007/s11103-009-9478-x URL |
| [13] |
Ibiza V P, Blanca J, Cañizares J, Nuez F. 2012. Taxonomy and genetic diversity of domesticated Capsicum species in the Andean region. Genetic Resources and Crop Evolution, 59(6):1077-1088.
doi: 10.1007/s10722-011-9744-z URL |
| [14] |
Jaeger K E, Wigge P A. 2007. FT protein acts as a long-range signal in Arabidopsis. Current Biology, 17(12):1050-1054.
pmid: 17540569 |
| [15] | Kaneko-Suzuki M, Kurihara-Ishikawa R, Okushita-Terakawa C, Kojima C, Nagano-Fujiwara M, Ohki I, Tsuji H, Shimamoto K, Taoka K I. 2018. TFL1-Like proteins in rice antagonize rice FT-Like protein in inflorescence development by competition for complex formation with 14-3-3 and FD. Plant & Cell Physiology, 59(3):458-468. |
| [16] |
Karlgren A, Gyllenstrand N, Källman T, Sundström J F, Moore D, Lascoux M, Lagercrantz U. 2011. Evolution of the PEBP gene family in plants:functional diversification in seed plant evolution. Plant Physiology, 156(4):1967-1977.
doi: 10.1104/pp.111.176206 URL |
| [17] |
Kim S, Park J, Yeom S I, Kim Y M, Seo E, Kim K T, Kim M S, Lee J M, Cheong K, Shin H S, Kim S B, Han K, Lee J, Park M, Lee H A, Lee H Y, Lee Y, Oh S, Lee J H, Choi E, Choi E, Lee S E, Jeon J, Kim H, Choi G, Song H, Lee J, Lee S C, Kwon J K, Lee H Y, Koo N, Hong Y, Kim R W, Kang W H, Huh J H, Kang B C, Yang T J, Lee Y H, Bennetzen J L, Choi D. 2017. New reference genome sequences of hot pepper reveal the massive evolution of plant disease-resistance genes by retroduplication. Genome Biology, 18(1):210.
doi: 10.1186/s13059-017-1341-9 URL |
| [18] |
Kim S, Park M, Yeom S I, Kim Y M, Lee J M, Lee H A, Seo E, Choi J, Cheong K, Kim K T, Jung K, Lee G W, Oh H K, Bae S Y, Kim S Y, Jo Y D, Yang H B, Kang W H, Yu Y, Bark B S, Kim R Y, Choi I K, Choi B S, Lim J S, Less Y H, Choi D. 2014. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nature Genetics, 46(3):270-279.
doi: 10.1038/ng.2877 URL |
| [19] |
Kumar S, Stecher G, Tamura K. 2016. MEGA7:molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33(7):1870-1874.
doi: 10.1093/molbev/msw054 URL |
| [20] |
Li Chao, Zhang Yan-nan, Liu Huan-long, Huang Xian-zhong. 2015. Identification of PEBP gene family in Gossypium arboreum and Gossypium raimondii and expression analysis of the gene family in Gossypium hirsutum . Acta Agronomica Sinica, 41(3):394-404. (in Chinese)
doi: 10.3724/SP.J.1006.2015.00394 URL |
| 李超, 张彦楠, 刘焕龙, 黄先忠. 2015. 亚洲棉和雷蒙德氏棉PEBP家族基因的鉴定及该家族基因在陆地棉组织中表达分析. 作物学报, 41(3):394-404. | |
| [21] |
Li Q, Fan C, Zhang X, Wang X, Wu F, Hu R, Fu Y. 2014. Identification of a soybean MOTHER OF FT AND TFL1 homolog involved in regulation of seed germination. PLoS ONE, 9(6):e99642.
doi: 10.1371/journal.pone.0099642 URL |
| [22] |
Lifschitz E, Eshed Y. 2006. Universal florigenic signals triggered by FT homologues regulate growth and flowering cycles in perennial day-neutral tomato. Journal of Experimental Botany, 57(13):3405-3414.
doi: 10.1093/jxb/erl106 URL |
| [23] |
Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods, 25(4):402-408.
pmid: 11846609 |
| [24] |
Molinero-Rosales N, Latorre A, Jamilena M, Lozano R. 2004. SINGLE FLOWER TRUSS regulates the transition and maintenance of flowering in tomato. Planta, 218(3):427-434.
pmid: 14504922 |
| [25] |
Nakamura S, Abe F, Kawahigashi H, Nakazono K, Tagiri A, Matsumoto T, Utsugi S, Ogawa T, Handa H, Ishida H, Mori M, Kawaura K, Ogihara Y, Miura H. 2011. A wheat homolog of MOTHER OF FT AND TFL1 acts in the regulation of germination. The Plant Cell, 23(9):3215-3229.
doi: 10.1105/tpc.111.088492 pmid: 21896881 |
| [26] |
Navarro C, Abelenda J A, Cruz-Oró E, Cuéllar C A, Tamaki S, Silva J, Shimamoto k, Prat S. 2011. Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature, 478(7367):119-122.
doi: 10.1038/nature10431 URL |
| [27] | Qi Shiming, Liang Yan. 2020. Advances in research of the SELF-PRUNING gene family and plant architecture regulatory functions in tomato . Acta Horticulturae Sinica, 47(9):1705-1714. (in Chinese) |
| 祁世明, 梁燕. 2020. 番茄SELF-PRUNING基因家族及株形调控功能研究进展. 园艺学报, 47(9):1705-1714. | |
| [28] | Qin C, Yu C, Shen Y, Fang X, Chen L, Min J, Cheng J, Zhao S, Xu M, Luo Y, Yang Y, Wu H, Wu Z, Mao L, Zhou H, Lin H, Tang X, Zhao M, Huang Z, Zhou A, Yao X, Cui J, Li W, Chen Z, Feng Y, Niu Y, Yang X, Li W, Cai H. 2014. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proceedings of the National Academy of Science of the United States of America, 111(14):5135-5140. |
| [29] |
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio J C, Guirao-Rico S, Librado P, Ramos-Onsins S E, Sánchez-Gracia A. 2017. DnaSP 6:DNA sequence polymorphism analysis of large datasets. Molecular Biology and Evolution, 34(12):3299-3302.
doi: 10.1093/molbev/msx248 URL |
| [30] |
Wan H, Yuan W, Ruan M, Ye Q, Wang R, Li Z, Zhou G, Yao Z, Zhao J, Liu S, Yang Y. 2011. Identification of reference genes for reverse transcription quantitative real-time PCR normalization in pepper(Capsicum annuumL.). Biochemical and Biophysical Research Communications, 416(1-2):24-30.
doi: 10.1016/j.bbrc.2011.10.105 URL |
| [31] | Wang Y, Tang H, DeBarry J D, Tan X, Li J, Wang X, Lee T H, Jin H, Marler B, Guo H, Kissinger J C, Paterson A H. 2012. MCScanX:a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Research, 40(7):49. |
| [32] |
Wang Y D, Chen G J, Lei J J, Cao B H, Chen C M. 2020. Identification and characterization of a LEA-like gene,CaMF5,specifically expressed in the anthers of male-fertileCapsicum annuum. Horticultural Plant Journal, 6(1):39-48.
doi: 10.1016/j.hpj.2019.07.004 URL |
| [33] |
Wickland D P, Hanzawa Y. 2015. TheFLOWERING LOCUS T/TERMINAL FLOWER 1 gene family:functional evolution and molecular mechanisms. Molecular Plant, 8(7):983-997.
doi: 10.1016/j.molp.2015.01.007 pmid: 25598141 |
| [34] |
Xi W, Liu C, Hou X, Yu H. 2010. MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling inArabidopsis. The Plant Cell, 22(6):1733-1748.
doi: 10.1105/tpc.109.073072 URL |
| [35] |
Yadav C B, Bonthala V S, Muthamilarasan M, Pandey G, Khan Y, Prasad M. 2015. Genome-wide development of transposable elements-based markers in foxtail millet and construction of an integrated database. DNA Research, 22(1):79-90.
doi: 10.1093/dnares/dsu039 URL |
| [1] | WANG Xiaochen, NIE Ziye, LIU Xianju, DUAN Wei, FAN Peige, and LIANG Zhenchang, . Effects of Abscisic Acid on Monoterpene Synthesis in‘Jingxiangyu’Grape Berries [J]. Acta Horticulturae Sinica, 2023, 50(2): 237-249. |
| [2] | ZHAI Hanhan, ZHAI Yujie, TIAN Yi, ZHANG Ye, YANG Li, WEN Zhiliang, CHEN Haijiang. Genome-wide Identification of Peach SAUR Gene Family and Characterization of PpSAUR5 Gene [J]. Acta Horticulturae Sinica, 2023, 50(1): 1-14. |
| [3] | ZHANG Qiuyue, LIU Changlai, YU Xiaojing, YANG Jiading, FENG Chaonian. Screening of Reference Genes for Differentially Expressed Genes in Pyrus betulaefolia Plant Under Salt Stress by qRT-PCR [J]. Acta Horticulturae Sinica, 2022, 49(7): 1557-1570. |
| [4] | LI Yamei, MA Fuli, ZHANG Shanqi, HUANG Jinqiu, CHEN Mengting, ZHOU Junyong, SUN Qibao, SUN Jun. Optimization of Jujube Callus Transformation System and Application of ZjBRC1 in Regulating ZjYUCCA Expression [J]. Acta Horticulturae Sinica, 2022, 49(4): 749-757. |
| [5] | WANG Ying, AI Penghui, LI Shuailei, KANG Dongru, LI Zhongai, WANG Zicheng. Identification and Expression Analysis of Genes Related to DNA Methylation in Chrysanthemum × morifolium and C. nankingense [J]. Acta Horticulturae Sinica, 2022, 49(4): 827-840. |
| [6] | ZHANG Rui, ZHANG Xiayi, ZHAO Ting, WANG Shuangcheng, ZHANG Zhongxing, LIU Bo, ZHANG De, WANG Yanxiu. Transcriptome Analysis of the Molecular Mechanism of Saline-alkali Stress Response in Malus halliana Leaves [J]. Acta Horticulturae Sinica, 2022, 49(2): 237-251. |
| [7] | ZHOU Zhiming, YANG Jiabao, ZHANG Cheng, ZENG Linglu, MENG Wanqiu, SUN Li. Genome-wide Identification and Expression Analyses of Long-chain Acyl-CoA Synthetases Under Abiotic Stresses in Helianthus annuus [J]. Acta Horticulturae Sinica, 2022, 49(2): 352-364. |
| [8] | QIAO Jun, WANG Liying, LIU Jing, LI Suweng. Expression Analysis of Genes Related to Photosensitive Color Under the Caylx in Eggplant Based on Transcriptome Sequencing [J]. Acta Horticulturae Sinica, 2022, 49(11): 2347-2356. |
| [9] | HOU Tianze, YI Shuangshuang, ZHANG Zhiqun, WANG Jian, LI Chonghui. Selection and Validation of Reference Genes for RT-qPCR in Phalaenopsis- type Dendrobium Hybrid [J]. Acta Horticulturae Sinica, 2022, 49(11): 2489-2501. |
| [10] | ZHOU Tie, PAN Bin, LI Feifei, MA Xiaochuan, TANG Mengjing, LIAN Xuefei, CHANG Yuanyuan, CHEN Yuewen, LU Xiaopeng. Effects of Drought Stress at Enlargement Stage on Fruit Quality Formation of Satsuma Mandarin and the Law of Water Absorption and Transportation in Tree After Re-watering [J]. Acta Horticulturae Sinica, 2022, 49(1): 11-22. |
| [11] | HE Yan, SUN Yanli, ZHAO Fangfang, DAI Hongjun. Effect of Exogenous Brassinolides Treatment on Sugar Metabolism of Merlot Grape Berries [J]. Acta Horticulturae Sinica, 2022, 49(1): 117-128. |
| [12] | LI Maofu, YANG Yuan, WANG Hua, FAN Youwei, SUN Pei, JIN Wanmei. Identification and Analysis of Self Incompatibility S-RNase in Rose [J]. Acta Horticulturae Sinica, 2022, 49(1): 157-165. |
| [13] | QI Xiliang, LIU Congli, SONG Lulu, LI Ming. Functional Analysis of Sucrose-phosphate Synthase Genes(SPS)in Sweet Cherry [J]. Acta Horticulturae Sinica, 2021, 48(8): 1446-1456. |
| [14] | YAO Fuwen, WANG Meige, SONG Chunhui, SONG Shangwei, JIAO Jian, WANG Miaomiao, WANG Kun, BAI Tuanhui, ZHENG Xianbo. Identification and Expression Analysis of HSP90 Gene Family Under High Temperature Stress in Apple [J]. Acta Horticulturae Sinica, 2021, 48(5): 849-859. |
| [15] | CHEN Zumin, XIAO Nuoya, ZHANG Yanxia, SHI Xiaomin, GUO Shuaiqi, GAO Hu, WANG Zhenping. Effects of Water Stress on the Volatile Compounds and Related Biosynthetic Genes Expression in‘Muscat Hamburg’Grape Berries [J]. Acta Horticulturae Sinica, 2021, 48(5): 883-896. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd