
Acta Horticulturae Sinica ›› 2021, Vol. 48 ›› Issue (3): 439-455.doi: 10.16420/j.issn.0513-353x.2020-0421
• Research Papers • Previous Articles Next Articles
HAN Wenlong1, ZHU Zhen1, LI Junru1, CHEN Tianzhe1, LI Guohui1, SU Xueqiang1, JIN Qin1,2, CHENG Xi1,2, CAI Yongping1,2,*( )
)
Received:2020-09-28
Online:2021-03-25
Published:2021-04-02
Contact:
CAI Yongping
E-mail:swkx12@ahau.edu.cn
CLC Number:
HAN Wenlong, ZHU Zhen, LI Junru, CHEN Tianzhe, LI Guohui, SU Xueqiang, JIN Qin, CHENG Xi, CAI Yongping. Indentification and Expression Analysis of Trihelix Transcription Factor Family in Pear[J]. Acta Horticulturae Sinica, 2021, 48(3): 439-455.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2020-0421
| 基因名称 Gene name | 上游引物(5′-3′) Forward primer | 下游引物(5′-3′) Reverse primer |
|---|---|---|
| PbGT1 | AGTGAACTGACTCGAGCGATATTGA | ACTAGCATTCCCAACCCCGTTAACC |
| PbGT2 | CACGGGTTGTTTTGTTTATTACCTT | TTGTTGCTGCTACGATGGTTACTAG |
| PbGT3 | AGATGATGGAGCTGGAGAAACAGAG | AATGATCAAATAAAACCCATAAACA |
| PbGT4 | GAATATGTTCATGGATGTTCAGGTG | TGCCTACATTGCAGTAATTCTTAAC |
| PbGT5 | AAACCAGTGGTGATGGGACTAAGTG | TGCGACTTCATGATTTTCCATTTTT |
| PbGT6 | AAAAGAGTAAAAGGAAGCAGATGGC | CAGATAGATACCGATATTACCCGCT |
| PbGT7 | TTCGTATAGAGTGTTGGCGGATTCA | ATCATTCACTGAGATCGTCAACAGA |
| PbGT8 | GTTGGAGTTGCAGAAGAAGCAGATT | AGATACCACACACCCTCACCTGCAT |
| PbGT9 | TGCCTCTATGACGAGTCGGACCACA | GGAACGCGGTTCCCTCAACTCACAC |
| PbGT10 | GCATCGGCAGCTTGTTACTTTCAGA | CACTGGTTTTGGCTTTCCGTGCTGA |
| PbGT11 | CTCCTAATGGTGGGAATAATGCAAG | ATCATCACTGGTTTTGGCTTCCCTT |
| PbGT12 | GAGGGACTTTCACCATTGTGTTCTA | TTCAAAAAATGCATCACCTGTCCAA |
| PbGT13 | AGAATACAGGCCGTTCGTCTAACGG | ATTATCTCATTGAACCATGGCGAGG |
| PbGT14 | AATAGGATGGAGGAAGCGGATAGCG | CACATAACTCACTGAACCATGGCGA |
| PbGT15 | TAAGAATGATGCTAGTTATGTGGAG | GCCATTCTATCTCACTCGTATCATG |
| PbGT16 | GAGGATTCAAAAACATGTCCATATT | TGATGACTCTTTCTAGTTGGTTACC |
| Tubulin | AGAACAAGAACTCGTCCTAC | GAACTGCTCGCTCACTCTCC |
Table 1 Primer sequences used in this study
| 基因名称 Gene name | 上游引物(5′-3′) Forward primer | 下游引物(5′-3′) Reverse primer |
|---|---|---|
| PbGT1 | AGTGAACTGACTCGAGCGATATTGA | ACTAGCATTCCCAACCCCGTTAACC |
| PbGT2 | CACGGGTTGTTTTGTTTATTACCTT | TTGTTGCTGCTACGATGGTTACTAG |
| PbGT3 | AGATGATGGAGCTGGAGAAACAGAG | AATGATCAAATAAAACCCATAAACA |
| PbGT4 | GAATATGTTCATGGATGTTCAGGTG | TGCCTACATTGCAGTAATTCTTAAC |
| PbGT5 | AAACCAGTGGTGATGGGACTAAGTG | TGCGACTTCATGATTTTCCATTTTT |
| PbGT6 | AAAAGAGTAAAAGGAAGCAGATGGC | CAGATAGATACCGATATTACCCGCT |
| PbGT7 | TTCGTATAGAGTGTTGGCGGATTCA | ATCATTCACTGAGATCGTCAACAGA |
| PbGT8 | GTTGGAGTTGCAGAAGAAGCAGATT | AGATACCACACACCCTCACCTGCAT |
| PbGT9 | TGCCTCTATGACGAGTCGGACCACA | GGAACGCGGTTCCCTCAACTCACAC |
| PbGT10 | GCATCGGCAGCTTGTTACTTTCAGA | CACTGGTTTTGGCTTTCCGTGCTGA |
| PbGT11 | CTCCTAATGGTGGGAATAATGCAAG | ATCATCACTGGTTTTGGCTTCCCTT |
| PbGT12 | GAGGGACTTTCACCATTGTGTTCTA | TTCAAAAAATGCATCACCTGTCCAA |
| PbGT13 | AGAATACAGGCCGTTCGTCTAACGG | ATTATCTCATTGAACCATGGCGAGG |
| PbGT14 | AATAGGATGGAGGAAGCGGATAGCG | CACATAACTCACTGAACCATGGCGA |
| PbGT15 | TAAGAATGATGCTAGTTATGTGGAG | GCCATTCTATCTCACTCGTATCATG |
| PbGT16 | GAGGATTCAAAAACATGTCCATATT | TGATGACTCTTTCTAGTTGGTTACC |
| Tubulin | AGAACAAGAACTCGTCCTAC | GAACTGCTCGCTCACTCTCC |
| 物种 Species | 基因编号 Gene ID | 基因名称 Gene name | 染色体 Chr | 蛋白质Protein | |||
|---|---|---|---|---|---|---|---|
| 等电点 pI | 长度/aa Length | 分子质量/kD Molecular weight | 亚细胞定位 Subcellular localization | ||||
| 梨 | Pbr022283.1 | PbGT1 | Chr17 | 8.34 | 585 | 63.32 | 细胞核 Nuclear |
| Pear | Pbr027539.1 | PbGT2 | Chr9 | 9.61 | 350 | 38.01 | 细胞核 Nuclear |
| Pbr009547.1 | PbGT3 | Chr1 | 9.27 | 247 | 27.90 | 细胞核 Nuclear | |
| Pbr010927.1 | PbGT4 | Chr7 | 9.20 | 241 | 27.34 | 细胞核 Nuclear | |
| Pbr016131.2 | PbGT5 | Chr10 | 5.50 | 448 | 49.17 | 细胞核 Nuclear | |
| Pbr034589.1 | PbGT6 | Chr5 | 4.70 | 398 | 45.36 | 细胞核 Nuclear | |
| Pbr027585.1 | PbGT7 | Chr2 | 4.79 | 363 | 42.82 | 细胞核 Nuclear | |
| Pbr015453.1 | PbGT8 | Chr15 | 4.90 | 360 | 42.54 | 细胞核 Nuclear | |
| Pbr036250.1 | PbGT9 | Chr16 | 5.95 | 426 | 48.87 | 细胞核 Nuclear | |
| Pbr016796.1 | PbGT10 | Chr14 | 6.40 | 532 | 60.38 | 细胞核 Nuclear | |
| Pbr005503.1 | PbGT11 | Chr12 | 6.36 | 536 | 60.58 | 细胞核 Nuclear | |
| Pbr027610.1 | PbGT12 | Chr7 | 5.76 | 566 | 65.15 | 细胞核 Nuclear | |
| Pbr038634.1 | PbGT13 | Chr6 | 5.65 | 758 | 82.46 | 细胞核 Nuclear | |
| Pbr010171.1 | PbGT14 | Chr14 | 5.57 | 647 | 71.99 | 细胞核 Nuclear | |
| Pbr010535.1 | PbGT15 | Chr5 | 6.60 | 572 | 64.69 | 细胞核 Nuclear | |
| Pbr036349.1 | PbGT16 | Chr10 | 8.73 | 534 | 61.13 | 细胞核 Nuclear | |
| 草莓 | mrna29666 | FvGT1 | LG3 | 5.00 | 318 | 35.19 | 细胞核 Nuclear |
| Strawberry | mrna21007 | FvGT2 | LG7 | 8.43 | 284 | 32.41 | 细胞核 Nuclear |
| mrna05400 | FvGT3 | LG6 | 9.78 | 378 | 40.84 | 细胞核 Nuclear | |
| mrna05956 | FvGT4 | LG6 | 4.68 | 395 | 45.27 | 细胞核 Nuclear | |
| mrna02091 | FvGT5 | LG1 | 4.87 | 364 | 42.27 | 细胞核 Nuclear | |
| mrna30101 | FvGT6 | LG6 | 6.81 | 591 | 64.60 | 细胞核 Nuclear | |
| mrna27722 | FvGT7 | LG5 | 6.55 | 376 | 43.32 | 细胞核 Nuclear | |
| mrna01828 | FvGT8 | LG5 | 5.56 | 769 | 83.55 | 细胞核 Nuclear | |
| mrna08173 | FvGT9 | LG2 | 6.21 | 578 | 65.76 | 细胞核 Nuclear | |
| mrna23617 | FvGT10 | LG7 | 6.19 | 433 | 49.00 | 细胞核 Nuclear | |
| mrna30156 | FvGT11 | LG6 | 6.42 | 579 | 65.77 | 细胞核 Nuclear | |
| 桃 | Prupe.4G061800.1 | PpGT1 | Pp4 | 5.21 | 310 | 34.69 | 细胞核 Nuclear |
| Peach | Prupe.3G118000.1 | PpGT2 | Pp3 | 9.88 | 326 | 37.02 | 细胞核 Nuclear |
| Prupe.2G253200.1 | PpGT3 | Pp2 | 9.38 | 548 | 61.78 | 叶绿体 Chloroplast | |
| Prupe.3G036000.1 | PpGT4 | Pp3 | 9.53 | 364 | 39.55 | 细胞核 Nuclear | |
| Prupe.3G071500.1 | PpGT5 | Pp3 | 4.59 | 398 | 45.72 | 细胞核 Nuclear | |
| Prupe.7G228600.1 | PpGT6 | Pp7 | 4.85 | 361 | 42.57 | 细胞核 Nuclear | |
| Prupe.7G120600.1 | PpGT7 | Pp7 | 6.41 | 527 | 57.10 | 细胞核 Nuclear | |
| Prupe.2G090800.1 | PpGT8 | Pp2 | 8.52 | 491 | 53.39 | 细胞核 Nuclear | |
| Prupe.1G235200.1 | PpGT9 | Pp1 | 5.90 | 380 | 43.75 | 细胞核 Nuclear | |
| Prupe.5G017700.1 | PpGT10 | Pp5 | 8.66 | 286 | 33.26 | 细胞核 Nuclear | |
| Prupe.7G116400.1 | PpGT11 | Pp7 | 6.40 | 544 | 61.86 | 细胞核 Nuclear | |
| Prupe.2G086500.1 | PpGT12 | Pp2 | 6.20 | 565 | 64.17 | 细胞核 Nuclear | |
| Prupe.5G137600.1 | PpGT13 | Pp5 | 5.26 | 776 | 84.46 | 细胞核 Nuclear | |
| Prupe.8G028800.1 | PpGT14 | Pp8 | 6.77 | 590 | 66.96 | 细胞核 Nuclear | |
| Prupe.8G028700.1 | PpGT15 | Pp8 | 6.05 | 630 | 70.57 | 细胞核 Nuclear | |
| Prupe.6G162600.1 | PpGT16 | Pp6 | 5.49 | 547 | 62.28 | 细胞核 Nuclear | |
Table 2 Information on Trihelix family members and physicochemical properties of GT protein of pear,strawberry and peach
| 物种 Species | 基因编号 Gene ID | 基因名称 Gene name | 染色体 Chr | 蛋白质Protein | |||
|---|---|---|---|---|---|---|---|
| 等电点 pI | 长度/aa Length | 分子质量/kD Molecular weight | 亚细胞定位 Subcellular localization | ||||
| 梨 | Pbr022283.1 | PbGT1 | Chr17 | 8.34 | 585 | 63.32 | 细胞核 Nuclear |
| Pear | Pbr027539.1 | PbGT2 | Chr9 | 9.61 | 350 | 38.01 | 细胞核 Nuclear |
| Pbr009547.1 | PbGT3 | Chr1 | 9.27 | 247 | 27.90 | 细胞核 Nuclear | |
| Pbr010927.1 | PbGT4 | Chr7 | 9.20 | 241 | 27.34 | 细胞核 Nuclear | |
| Pbr016131.2 | PbGT5 | Chr10 | 5.50 | 448 | 49.17 | 细胞核 Nuclear | |
| Pbr034589.1 | PbGT6 | Chr5 | 4.70 | 398 | 45.36 | 细胞核 Nuclear | |
| Pbr027585.1 | PbGT7 | Chr2 | 4.79 | 363 | 42.82 | 细胞核 Nuclear | |
| Pbr015453.1 | PbGT8 | Chr15 | 4.90 | 360 | 42.54 | 细胞核 Nuclear | |
| Pbr036250.1 | PbGT9 | Chr16 | 5.95 | 426 | 48.87 | 细胞核 Nuclear | |
| Pbr016796.1 | PbGT10 | Chr14 | 6.40 | 532 | 60.38 | 细胞核 Nuclear | |
| Pbr005503.1 | PbGT11 | Chr12 | 6.36 | 536 | 60.58 | 细胞核 Nuclear | |
| Pbr027610.1 | PbGT12 | Chr7 | 5.76 | 566 | 65.15 | 细胞核 Nuclear | |
| Pbr038634.1 | PbGT13 | Chr6 | 5.65 | 758 | 82.46 | 细胞核 Nuclear | |
| Pbr010171.1 | PbGT14 | Chr14 | 5.57 | 647 | 71.99 | 细胞核 Nuclear | |
| Pbr010535.1 | PbGT15 | Chr5 | 6.60 | 572 | 64.69 | 细胞核 Nuclear | |
| Pbr036349.1 | PbGT16 | Chr10 | 8.73 | 534 | 61.13 | 细胞核 Nuclear | |
| 草莓 | mrna29666 | FvGT1 | LG3 | 5.00 | 318 | 35.19 | 细胞核 Nuclear |
| Strawberry | mrna21007 | FvGT2 | LG7 | 8.43 | 284 | 32.41 | 细胞核 Nuclear |
| mrna05400 | FvGT3 | LG6 | 9.78 | 378 | 40.84 | 细胞核 Nuclear | |
| mrna05956 | FvGT4 | LG6 | 4.68 | 395 | 45.27 | 细胞核 Nuclear | |
| mrna02091 | FvGT5 | LG1 | 4.87 | 364 | 42.27 | 细胞核 Nuclear | |
| mrna30101 | FvGT6 | LG6 | 6.81 | 591 | 64.60 | 细胞核 Nuclear | |
| mrna27722 | FvGT7 | LG5 | 6.55 | 376 | 43.32 | 细胞核 Nuclear | |
| mrna01828 | FvGT8 | LG5 | 5.56 | 769 | 83.55 | 细胞核 Nuclear | |
| mrna08173 | FvGT9 | LG2 | 6.21 | 578 | 65.76 | 细胞核 Nuclear | |
| mrna23617 | FvGT10 | LG7 | 6.19 | 433 | 49.00 | 细胞核 Nuclear | |
| mrna30156 | FvGT11 | LG6 | 6.42 | 579 | 65.77 | 细胞核 Nuclear | |
| 桃 | Prupe.4G061800.1 | PpGT1 | Pp4 | 5.21 | 310 | 34.69 | 细胞核 Nuclear |
| Peach | Prupe.3G118000.1 | PpGT2 | Pp3 | 9.88 | 326 | 37.02 | 细胞核 Nuclear |
| Prupe.2G253200.1 | PpGT3 | Pp2 | 9.38 | 548 | 61.78 | 叶绿体 Chloroplast | |
| Prupe.3G036000.1 | PpGT4 | Pp3 | 9.53 | 364 | 39.55 | 细胞核 Nuclear | |
| Prupe.3G071500.1 | PpGT5 | Pp3 | 4.59 | 398 | 45.72 | 细胞核 Nuclear | |
| Prupe.7G228600.1 | PpGT6 | Pp7 | 4.85 | 361 | 42.57 | 细胞核 Nuclear | |
| Prupe.7G120600.1 | PpGT7 | Pp7 | 6.41 | 527 | 57.10 | 细胞核 Nuclear | |
| Prupe.2G090800.1 | PpGT8 | Pp2 | 8.52 | 491 | 53.39 | 细胞核 Nuclear | |
| Prupe.1G235200.1 | PpGT9 | Pp1 | 5.90 | 380 | 43.75 | 细胞核 Nuclear | |
| Prupe.5G017700.1 | PpGT10 | Pp5 | 8.66 | 286 | 33.26 | 细胞核 Nuclear | |
| Prupe.7G116400.1 | PpGT11 | Pp7 | 6.40 | 544 | 61.86 | 细胞核 Nuclear | |
| Prupe.2G086500.1 | PpGT12 | Pp2 | 6.20 | 565 | 64.17 | 细胞核 Nuclear | |
| Prupe.5G137600.1 | PpGT13 | Pp5 | 5.26 | 776 | 84.46 | 细胞核 Nuclear | |
| Prupe.8G028800.1 | PpGT14 | Pp8 | 6.77 | 590 | 66.96 | 细胞核 Nuclear | |
| Prupe.8G028700.1 | PpGT15 | Pp8 | 6.05 | 630 | 70.57 | 细胞核 Nuclear | |
| Prupe.6G162600.1 | PpGT16 | Pp6 | 5.49 | 547 | 62.28 | 细胞核 Nuclear | |
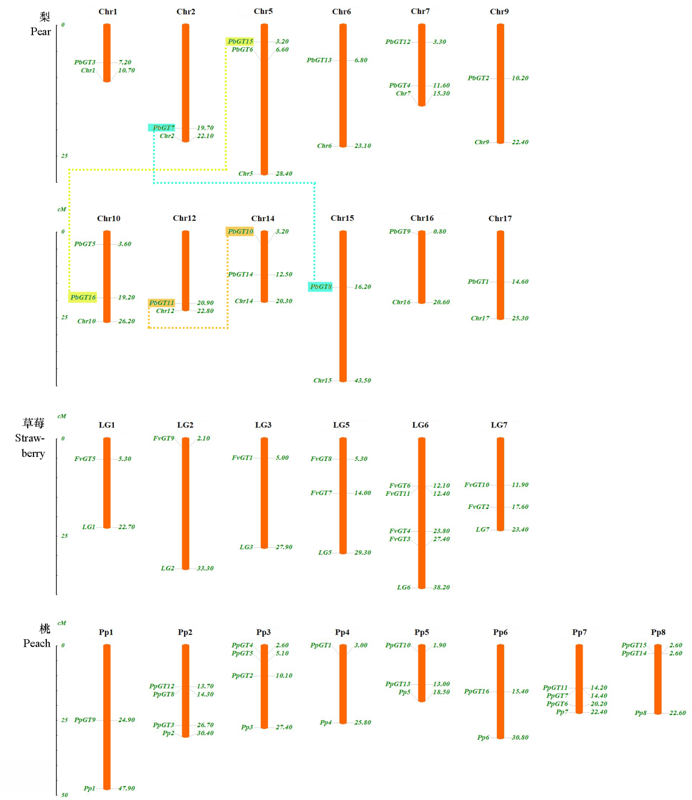
Fig. 2 Chromosome localization and gene duplication events analysis of the Trihelix family members of pear, strawberry and peach The two genes connected by dotted lines are segmental duplication.
| 复制基因对 Duplicated gene pairs | 非同义替换率 Ka | 同义替换率 Ks | 非同义替换率/同义替换率 Ka/Ks | 复制方式 Duplicated type |
|---|---|---|---|---|
| PbGT7/PbGT8 | 0.0333 | 0.1014 | 0.3284 | 片段复制 Segmental duplication |
| PbGT10/PbGT11 | 0.0329 | 0.1718 | 0.1915 | 片段复制 Segmental duplication |
| PbGT15/PbGT16 | 0.0646 | 0.1946 | 0.3320 | 片段复制 Segmental duplication |
Table 3 Gene duplication events analysis of the Trihelix family members of pear
| 复制基因对 Duplicated gene pairs | 非同义替换率 Ka | 同义替换率 Ks | 非同义替换率/同义替换率 Ka/Ks | 复制方式 Duplicated type |
|---|---|---|---|---|
| PbGT7/PbGT8 | 0.0333 | 0.1014 | 0.3284 | 片段复制 Segmental duplication |
| PbGT10/PbGT11 | 0.0329 | 0.1718 | 0.1915 | 片段复制 Segmental duplication |
| PbGT15/PbGT16 | 0.0646 | 0.1946 | 0.3320 | 片段复制 Segmental duplication |
| 基因名称 Gene name | ABRE | CGTCA motif | ERE | LTR | MBS | MRE | TGA-element | TC-rich repeats | AC-Ⅰ | AC-Ⅱ |
|---|---|---|---|---|---|---|---|---|---|---|
| PbGT1 | 1 | 3 | 1 | 2 | 2 | |||||
| PbGT2 | 5 | 7 | 1 | 1 | 1 | |||||
| PbGT3 | 9 | 5 | 1 | |||||||
| PbGT4 | 5 | 4 | 1 | 1 | 2 | 1 | 3 | 1 | ||
| PbGT5 | 6 | |||||||||
| PbGT6 | 1 | 2 | 3 | 1 | ||||||
| PbGT7 | 3 | 5 | 1 | 2 | 1 | 1 | 1 | |||
| PbGT8 | 4 | 2 | 1 | 1 | ||||||
| PbGT9 | 2 | 2 | ||||||||
| PbGT10 | 4 | 4 | 1 | 1 | 1 | |||||
| PbGT11 | 3 | 5 | 2 | 2 | ||||||
| PbGT12 | 4 | 2 | 1 | 1 | 2 | 1 | ||||
| PbGT13 | 1 | 1 | 1 | 1 | ||||||
| PbGT14 | 1 | 1 | 1 | |||||||
| PbGT15 | 3 | 1 | 1 | 1 | 1 | |||||
| PbGT16 | 3 | 5 | 1 | 1 | 1 |
Table 4 Promoter cis-acting elements prediction of the Trihelix family members of pear
| 基因名称 Gene name | ABRE | CGTCA motif | ERE | LTR | MBS | MRE | TGA-element | TC-rich repeats | AC-Ⅰ | AC-Ⅱ |
|---|---|---|---|---|---|---|---|---|---|---|
| PbGT1 | 1 | 3 | 1 | 2 | 2 | |||||
| PbGT2 | 5 | 7 | 1 | 1 | 1 | |||||
| PbGT3 | 9 | 5 | 1 | |||||||
| PbGT4 | 5 | 4 | 1 | 1 | 2 | 1 | 3 | 1 | ||
| PbGT5 | 6 | |||||||||
| PbGT6 | 1 | 2 | 3 | 1 | ||||||
| PbGT7 | 3 | 5 | 1 | 2 | 1 | 1 | 1 | |||
| PbGT8 | 4 | 2 | 1 | 1 | ||||||
| PbGT9 | 2 | 2 | ||||||||
| PbGT10 | 4 | 4 | 1 | 1 | 1 | |||||
| PbGT11 | 3 | 5 | 2 | 2 | ||||||
| PbGT12 | 4 | 2 | 1 | 1 | 2 | 1 | ||||
| PbGT13 | 1 | 1 | 1 | 1 | ||||||
| PbGT14 | 1 | 1 | 1 | |||||||
| PbGT15 | 3 | 1 | 1 | 1 | 1 | |||||
| PbGT16 | 3 | 5 | 1 | 1 | 1 |
| [1] |
Bremer B, Bremer K, Chase M W, Fay M F, Reveal J L, Soltis D E, Soltis P S, Stevens P F, Anderberg A A, Moore M J, Olmstead R G, Rudall P J, Sytsma K J, Tank D C, Wurdack K, Xiang J Q Y, Zmarzty S. 2009. An update of the angiosperm phylogeny group classification for the orders and families of flowering plants:APG Ⅲ. Botanical Journal of the Linnean Society, 161 (2):105-121.
doi: 10.1111/(ISSN)1095-8339 URL |
| [2] |
Brewer P B, Howles P A, Dorian K, Griffith M E, Ishida T, KaplanLevy R N, Kilinc A, Smyth D R. 2004. PETAL LOSS,a Trihelix transcription factor gene,regulates perianth architecture in the Arabidopsis flower. Development, 131 (16):4035-4045.
doi: 10.1242/dev.01279 URL |
| [3] |
Cai Yong-ping, Li Guo-hui, Nie Jing-quan, Lin Yi, Nie Fan, Zhang Jin-yun, Xu Yi-liu. 2010. Study of the structure and biosynthetic pathway of lignin in stone cells of pear. Scientia Horticulturae, 125 (3):374-379.
doi: 10.1016/j.scienta.2010.04.029 URL |
| [4] | Cheng Xi, Cai Yong-ping, Zhang Jin-yun. 2019. Stone cell development in pear//Schuyler S Korban. The pear genome. Compendium of Plant Genomes. Berlin: Springer Nature:201-225. |
| [5] |
Du He-wei, Huang Min, Liu Lei. 2016. The genome wide analysis of GT transcription factors that respond to drought and waterlogging stresses in maize. Euphytica, 208 (1):113-122.
doi: 10.1007/s10681-015-1599-5 URL |
| [6] |
Fan Ming-zhu, Herburger K, Jensen J K, Zemelis D S, Brandizzi F, Fry S C, Wilkerson C G. 2018. A Trihelix family transcription factor is associated with key genes in mixed-linkage glucan accumulation. Plant Physiology, 178 (3):1207-1221.
doi: 10.1104/pp.18.00978 URL |
| [7] |
Fang Yu-jie, Xie Ka-bin, Hou Xin, Hu Hong-hong, Xiong Li-zhong. 2010. Systematic analysis of GT factor family of rice reveals a novel subfamily involved in stress responses. Molecular Genetics and Genomics, 283 (2):157-169.
doi: 10.1007/s00438-009-0507-x URL |
| [8] |
Gao Hong-yan, Huang Rong, Liu Jun, Gao Zhi-min, Zhao Han-sheng, Li Xue-ping. 2019. Genome-wide identification of Trihelix genes in Moso Bamboo( Phyllostachys edulis)and their expression in response to abiotic stress. Journal of Plant Growth Regulation, 38 (3):1127-1140.
doi: 10.1007/s00344-019-09918-9 URL |
| [9] |
Green P J, Kay S A, Chua N H. 1987. Sequence-specific interactions of a pea nuclear factor with light-responsive elements upstream of the rbcS-3A gene. The EMBO Journal, 6 (9):2543-2549.
doi: 10.1002/embj.1987.6.issue-9 URL |
| [10] |
Han Li-bo, Li Yuan-bao, Wang Hai-yun, Wu Xiao-min, Li Chun-li, Luo Ming, Wu Shen-jie, Kong Zhao-sheng, Pei Yan, Jiao Gai-li, Xia Gui-xian. 2013. The dual functions of WLIM1a in cell elongation and secondary wall formation in developing cotton fibers. The Plant Cell, 25 (11):4421-4438.
doi: 10.1105/tpc.113.116970 URL |
| [11] |
Imagawa M, Sakaue R, Tanabe A, Osada S, Nishihara T. 2000. Two nuclear localization signals are required for nuclear translocation of nuclear factor 1-A. FEBS Letters, 484 (2):118-124.
doi: 10.1016/S0014-5793(00)02119-0 URL |
| [12] |
Jeroen R, Antje R, Jørgen H C, Yves V P, Wout B. 2003. Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiology, 133 (3):1051-1071.
doi: 10.1104/pp.103.026484 URL |
| [13] | Ji Jian-hui, Zhou Ying-jun, Wu He-he, Yang Li-ming. 2015. Genome-wide analysis and functional prediction of the Trihelix transcription factor family in rice. Herditas, 37 (12):1228-1241. (in Chinese) |
| 纪剑辉, 周颖君, 吴贺贺, 杨立明. 2015. 水稻Trihelix转录因子家族全基因组分析及功能预测. 遗传, 37 (12):1228-1241. | |
| [14] | Ji Qiao-hua, Xue Bao-ping, Zhang Xin-cao, Zhang Hao, Dang Feng-feng, Lin Jin-hui, He Wei-hong, Wang Yan-feng. 2018. Bioinformatics analysis of Trihelix transcription factors from Ziziphus jujuba. Molecular Plant Breeding, 16 (21):6968-6974. (in Chinese) |
| 姬俏华, 薛宝平, 张新草, 张昊, 党峰峰, 林金辉, 何伟红, 王延峰. 2018. 枣Trihelix转录因子家族生物信息学分析. 分子植物育种, 16 (21):6968-6974. | |
| [15] |
Kong Xiang-pai, Lü Wei, Jiang Shan-shan, Zhang Dan, Cai Guo-hua, Pan Jiao-wen, Li De-quan. 2013. Genome-wide identification and expression analysis of calcium-dependent protein kinase in maize. BMC Genomics, 14:433.
doi: 10.1186/1471-2164-14-433 URL |
| [16] |
Lampugnani E R, Kilinc A, Smyth D R. 2012. PETAL LOSS is a boundary gene that inhibits growth between developing sepals in Arabidopsis thaliana. Plant Journal, 71 (5):724-735.
doi: 10.1111/tpj.2012.71.issue-5 URL |
| [17] |
Li Guo-hui, Wang Han, Cheng Xi, Su Xue-qiang, Zhao Yu, Jiang Tao-shan. 2019. Comparative genomic analysis of the PAL genes in five Rosaceae species and functional identification of Chinese white pear. PeerJ, 7:e8064. doi: 10.7717/peerj.8064.
doi: 10.7717/peerj.8064 URL |
| [18] |
Li Jia-ming, Zhang Ming-hui, Sun Jian, Mao Xin-rui, Wang Jing, Wang Jing-guo, Liu Hua-long, Zheng Hong-liang, Zhen Zhen, Zhao Hong-wei, Zou De-tang. 2019. Genome-wide characterization and identification of Trihelix transcription factor and expression profiling in response to abiotic stresses in rice( Oryza sativa L.). International Journal of Molecular Sciences, 20 (2):251.
doi: 10.3390/ijms20020251 URL |
| [19] | Liu Xiao-yang, Li Ling, Zong Mei, Cai Yong-ping. 2004. Contents and distributions of stone cell and their effects on fruit quality of pear. Journal of Anhui Agricultural University, 31 (1):104-106. (in Chinese) |
| 刘小阳, 李玲, 宗梅, 蔡永萍. 2004. 梨果实石细胞含量分布及其对梨品质的影响. 安徽农业大学学报, 31 (1):104-106. | |
| [20] | Li Yue, Sun Jie, Xie Zong-ming, Li Quan-sheng, Si Ai-jun, Chen Shou-yi. 2013. Expression profiling of cotton( Gossypium hirsutum L.)Trihelix genes responsive to abiotic stresses. Scientia Agricultura Sinica, 46 (9):1946-1955. (in Chinese) |
| 李月, 孙杰, 谢宗铭, 李全胜, 司爱君, 陈受宜. 2013. 陆地棉Trihelix转录因子基因响应非生物胁迫表达谱分析. 中国农业科学, 46 (9):1946-1955. | |
| [21] |
Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2 -ΔΔCT method. Methods, 25 (4):402-408.
pmid: 11846609 |
| [22] | Lu Shi-xiong, Wang Ping, He Hong-hong, Liang Guo-ping, Ma Zong-huan, Qiao Ya-li, Wu Yu-xia, Chen Bai-hong, Mao Juan. 2019. Bioinformatics identification and expression analysis of grape Trihelix transcription factor family. Acta Horticulturae Sinica, 46 (7):1257-1269. (in Chinese) |
| 卢世雄, 王萍, 何红红, 梁国平, 马宗桓, 乔亚丽, 吴玉霞, 陈佰鸿, 毛娟. 2019. 葡萄Trihelix转录因子家族生物信息及其基因表达分析. 园艺学报, 46 (7):1257-1269. | |
| [23] |
Ma Zhao-tang, Liu Mo-yan, Sun Wen-jun, Huang Li, Wu Qi, Bu Tong-liang, Li Cheng-lei, Chen Hui. 2019. Genome-wide identification and expression analysis of the Trihelix transcription factor family in tartary buckwheat (Fagopyrum tataricum). BMC Plant Biology, 19 (1):344.
doi: 10.1186/s12870-019-1957-x URL |
| [24] |
Mo Hui-juan, Wang Ling-ling, Ma Shu-ya, Yue Dao-qian, Lu Li-li, Yang Zhao-en, Yang Zuo-ren, Li Fu-guang. 2019. Transcriptome profiling of Gossypium arboreum during fiber initiation and the genome-wide identification of Trihelix transcription factors. Gene, 709:36-47.
doi: 10.1016/j.gene.2019.02.091 URL |
| [25] |
Ouni R, Anna Zborowska A, Sehic J, Choulak S, Iñaki H J, Garkava-Gustavsson L, Mars M. 2020. Genetic diversity and structure of Tunisian local pear germplasm as revealed by SSR markers. Horticultural Plant Journal, 6 (2):61-70.
doi: 10.1016/j.hpj.2020.03.003 URL |
| [26] | Praveen G, Vineet K. 2020. Plant small RNA biogenesis,regulation and application. London:Elsevier:469-483. |
| [27] |
Riechmann J L, Heard J, Martin G, Reuber L, Jiang C Z, Keddie J, Adam L, Pineda O, Ratcliffe O J, Samaha R R, Creelman R, Pilgrim M, Broun P, Zhang J Z, Ghandehari D, Sherman B K, Yu G L. 2000. Arabidopsis transcription factors:genome-wide comparative analysis among eukaryotes. Science, 290 (5499):2105-2110.
doi: 10.1126/science.290.5499.2105 URL |
| [28] |
Su Xue-qiang, Zhao Yu, Wang Han, Li Guo-hui, Cheng Xi, Jin Qing, Cai Yong-ping. 2019. Transcriptomic analysis of early fruit development in Chinese white pear( Pyrus bretschneideri Rehd.)and functional identification of PbCCR1 in lignin biosynthesis. BMC Plant Biology, 19 (1):417.
doi: 10.1186/s12870-019-2046-x URL |
| [29] | Tian Lu-ming, Dong Xing-guang, Cao Yu-fen, Zhang Ying, Qi Dan. 2017. Correlation of flesh in Pyrus fruit with its stone cells lignin. Southwest China Journal of Agricultural Sciences, 30 (9):2091-2096. (in Chinese) |
| 田路明, 董星光, 曹玉芬, 张莹, 齐丹. 2017. 梨属植物果肉和石细胞团木质素相关性分析. 西南农业学报, 30 (9):2091-2096. | |
| [30] | Wang Bin, Zhang Nan, Yan Chong-chong, Jin Qing, Lin Yi, Cai Yong-ping, Zhang Jin-yun. 2013. Bagging for the development of stone cell and metabolism of lignin in Pyrus bretschneideri‘Dangshan Suli’. Acta Horticulturae Sinica, 40 (3):531-539. (in Chinese) |
| 王斌, 张楠, 闫冲冲, 金青, 林毅, 蔡永萍, 张金云. 2013. 套袋对砀山酥梨果实石细胞发育及木质素代谢的影响. 园艺学报, 40 (3):531-539. | |
| [31] |
Wang Huan-zhong, Yang J H, Chen Fang, Torres-Jerez I, Tang Yu-hong, Wang Ming-yi, Du Qian, Cheng Xiao-feng, Wen Jiang-qi, Dixon R. 2016. Transcriptome analysis of secondary cell wall development in Medicago truncatula. BMC Genomics, 17:23.
doi: 10.1186/s12864-015-2330-6 URL |
| [32] | Wang Ping, Lu Shi-xiong, Liang Guo-ping, Ma Zong-huan, Li Wen-fang, Mao Juan, Chen Bai-hong. 2019. Bioinformatics identification and expression analysis of Trihelix transcription factor family in apple. Acta Horticulturae Sinica, 46 (11):2082-2098. (in Chinese) |
| 王萍, 卢世雄, 梁国平, 马宗桓, 李文芳, 毛娟, 陈佰鸿. 2019. 苹果Trihelix转录因子家族生物信息学鉴定与基因表达分析. 园艺学报, 46 (11):2082-2098. | |
| [33] |
Wang Zhen-yi, Zhao Kang-lu, Pan Yu-xin, Wang Jin-peng, Song Xiao-ming, Ge Wei-na, Yuan Min, Lei Tian-yu, Wang Li, Zhang Lan, Li Yu-xian, Liu Tao, Chen Wei, Meng Wen-jing, Sun Chang-kai, Cui Xiao-bao, Bai Yun, Wang Xi-yin. 2018. Genomic,expressional,protein-protein interactional analysis of Trihelix transcription factor genes in Setaria italia and inference of their evolutionary trajectory. BMC Genomics, 19:665.
doi: 10.1186/s12864-018-5051-9 URL |
| [34] | Wei Chao-ling, Yang Hua, Wang Song-bo, Zhao Jian, Liu Chun, Gao Li-ping, Xia En-hua, Lu Ying, Tai Yu-ling, She Guang-biao, Sun Jun, Cao Hai-sheng, Tong Wei, Gao Qiang, Li Ye-yun, Deng Wei-wei, Jiang Xiao-lan, Wang Wen-zhao, Chen Qi, Zhang Shi-hua, Li Hai-jing, Wu Jun-lan, Wang Ping, Li Peng-hui, Shi Cheng-ying, Zheng Feng-ya, Jian Jian-bo, Huang Bei, Shan Dai, Shi Ming-ming, Fang Cong-bing, Yue Yi, Li Fang-dong, Li Da-xiang, Wei Shu, Han Bin, Jiang Chang-jun, Yin Ye, Xia Tao, Zhang Zheng-zhu, Bennetzen J L, Zhao Shan-cen, Wan Xiao-chun. 2018. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proceedings of the National Academy of Sciences of the United States of America, 115 (18):E4151-E4158. |
| [35] | Wu Tao, Zhang Rui-ping, Gu Chao, Wu Ju-you, Wan Hong-jian, Zhang Shu-jun, Zhang Shao-ling. 2012. Evaluation of candidate reference genes for real time quantitative PCR normalization in pear fruit. African Journal of Agricultural Research, 7 (25):3701-3704. |
| [36] | Xi Jing, Qiu Yong-jian, Du Li-qun, Poovaiah B W. 2012. Plant-specific Trihelix transcription factor AtGT2L interacts with calcium/calmodulin and responds to cold and salt stresses. Plant Science, 185:274-280. |
| [37] |
Yang Si-hai, Zhang Xiao-hui, Yue Jia-xing, Tian Da-cheng, Chen Jian-qun. 2008. Recent duplications dominate NBS-encoding gene expansion in two woody species. Molecular Genetics and Genomics, 280 (3):187-198.
doi: 10.1007/s00438-008-0355-0 URL |
| [38] |
Yu Chu-ying, Cai Xiao-feng, Ye Zhi-biao, Li Han-xia. 2015. Genome-wide identification and expression profiling analysis of Trihelix gene family in tomato. Biochemical and Biophysical Research Communications, 468 (4):653-659.
doi: 10.1016/j.bbrc.2015.11.010 URL |
| [39] | Yu Juan-juan, Li Ling, Jin Qing, Cai Yong-ping, Lin Yi, Lü Rui-hong. 2011. Studies on key enzyme POD types of lignin metabolic pathway during stone cell development of Pyrus bretschneideri. Acta Horticulturae Sinica, 38 (6):1037-1044. (in Chinese) |
| 于娟娟, 李玲, 金青, 蔡永萍, 林毅, 吕芮宏. 2011. 砀山酥梨石细胞发育过程中木质素代谢关键酶POD类型的分析. 园艺学报, 38 (6):1037-1044. | |
| [40] | Zhou Hong, Qian Jiao, Li Rong-fang, Chen Dan-dan, Fang Rong-jun, Zhao Wei-guo. 2017. Research of the Arabidopsis Trihelix transcription factor gene family. Journal of Jiangsu University of Science and Technology(Natural Science Edition), 31 (2):231-236. (in Chinese) |
| 周宏, 钱娇, 李荣芳, 陈丹丹, 方荣俊, 赵卫国. 2017. 拟南芥Trihelix转录因子基因家族研究. 江苏科技大学学报(自然科学版), 31 (2):231-236. |
| [1] | ZHAO Xueyan, WANG Qi, WANG Li, WANG Fangyuan, WANG Qing, LI Yan. Comparative Transcriptome Analysis of Differential Expression in Different Tissues of Corydalis yanhusuo [J]. Acta Horticulturae Sinica, 2023, 50(1): 177-187. |
| [2] | SONG Jiankun, YANG Yingjie, LI Dingli, MA Chunhui, WANG Caihong, and WANG Ran. A New Pear Cultivar‘Luxiu’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 3-4. |
| [3] | DONG Xingguang, CAO Yufen, ZHANG Ying, TIAN Luming, HUO Hongliang, QI Dan, XU Jiayu, LIU Chao, and WANG Lidong. A New Cold-resistant Crispy Pear Cultivar‘Yucuixiang’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 5-6. |
| [4] | OU Chunqing, JIANG Shuling, WANG Fei, MA Li, ZHANG Yanjie, and LIU Zhenjie. A New Early-ripening Pear Cultivar‘Xingli Mishui’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 7-8. |
| [5] | ZHANG Yanjie, WANG Fei, OU Chunqing, MA Li, JIANG Shuling, and LIU Zhenjie. A New Pear Cultivar‘Zhongli Yucui 3’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 9-10. |
| [6] | WANG Suke, LI Xiugen, YANG Jian, WANG Long, SU Yanli, ZHANG Xiangzhan, and XUE Huabai. A New Red Pear Cultivar‘Danxiahong’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 13-14. |
| [7] | WANG Fei, OU Chunqing, ZHANG Yanjie, MA Li, and JIANG Shuling. A New Late Ripening Pear Cultivar‘Huaqiu’with Long Storage Period [J]. Acta Horticulturae Sinica, 2022, 49(S1): 9-10. |
| [8] | SONG Jiankun, LI Dingli, YANG Yingjie, MA Chunhui, WANG Caihong, and WANG Ran. A New Pear Cultivar‘Qindaohong’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 11-12. |
| [9] | GUO Weizhen, ZHAO Jingxian, LI Ying. A New Mid-early Ripening Pear Cultivar‘Meiyu’ [J]. Acta Horticulturae Sinica, 2022, 49(9): 2051-2052. |
| [10] | GAO Yanlong, WU Yuxia, ZHANG Zhongxing, WANG Shuangcheng, ZHANG Rui, ZHANG De, WANG Yanxiu. Bioinformatics Analysis of Apple ELO Gene Family and Its Expression Analysis Under Low Temperature Stress [J]. Acta Horticulturae Sinica, 2022, 49(8): 1621-1636. |
| [11] | LIU Jinming, GUO Caihua, YUAN Xing, KANG Chao, QUAN Shaowen, NIU Jianxin. Genome-wide Identification of Dof Family Genes and Expression Analysis Sepal Persistent and Abscission in Pear [J]. Acta Horticulturae Sinica, 2022, 49(8): 1637-1649. |
| [12] | QIU Ziwen, LIU Linmin, LIN Yongsheng, LIN Xiaojie, LI Yongyu, WU Shaohua, YANG Chao. Cloning and Functional Analysis of the MbEGS Gene from Melaleuca bracteata [J]. Acta Horticulturae Sinica, 2022, 49(8): 1747-1760. |
| [13] | TAO Xin, ZHU Rongxiang, GONG Xin, WU Lei, ZHANG Shaoling, ZHAO Jianrong, ZHANG Huping. Fructokinase Gene PpyFRK5 Plays an Important Role in Sucrose Accumulation of Pear Fruit [J]. Acta Horticulturae Sinica, 2022, 49(7): 1429-1440. |
| [14] | ZHENG Lin, WANG Shuai, LIU Yunuo, DU Meixia, PENG Aihong, HE Yongrui, CHEN Shanchun, ZOU Xiuping. Gene Cloning and Expression Analysis of NAC Gene in Citrus in Response to Huanglongbing [J]. Acta Horticulturae Sinica, 2022, 49(7): 1441-1457. |
| [15] | MA Weifeng, LI Yanmei, MA Zonghuan, CHEN Baihong, MAO Juan. Identification of Apple POD Gene Family and Functional Analysis of MdPOD15 Gene [J]. Acta Horticulturae Sinica, 2022, 49(6): 1181-1199. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd