
Acta Horticulturae Sinica ›› 2021, Vol. 48 ›› Issue (2): 205-218.doi: 10.16420/j.issn.0513-353x.2020-0272
• Research Papers • Next Articles
YANG Yaming, DING Yuduan, CHEN Lijuan, TIAN Xueting, YIN Weijie, PENG Honghui, DU Wei, LIANG Liping, REN Xiaolin*( )
)
Received:2020-09-07
Revised:2020-11-11
Online:2021-02-25
Published:2021-03-09
Contact:
REN Xiaolin
E-mail:renxl@nwsuaf.edu.cn
CLC Number:
YANG Yaming, DING Yuduan, CHEN Lijuan, TIAN Xueting, YIN Weijie, PENG Honghui, DU Wei, LIANG Liping, REN Xiaolin. Identification and Expression Analysis of the Vacuolar Iron Transporter Gene Family in Apple[J]. Acta Horticulturae Sinica, 2021, 48(2): 205-218.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2020-0272
| 用途Usage | 基因Gene | 上游引物(5′-3′)Forward primer | 下游引物(5′-3′)Reverse primer |
|---|---|---|---|
| qRT-PCR | MdVIT1 | CTCCAATCCCCGAAGACGA | GCGATGCAACGGAAACCA |
| MdVIT2 | TCCGCTTCTCCCGTTAATATT | CGAGGTCGTTTGTGGTCATT | |
| MdVIT3 | CACCAAACAACGCTAGACCAA | TCACAGCTCCGACTCCCAT | |
| MdVIT4 | TTCGATTACTCCCAGAGGACAC | AGCGATGCAACAGAAACCAG | |
| MdVIT5 | AATCCCAGAAGGCGACGAA | GAGCGATGCAACAGAAACCAGCCCA | |
| MdVIT6 | TCTTACCGTGCCGTTTGCT | ATATCCGCCGAGTCCCATG | |
| MdVIT7 | CAAGGACCGAGCAAGTGCA | CATCAAGGACGTGGTTGACAGT | |
| MdVIT8 | CGACGGTCTCACTGTCCCTT | GGCGGCAACTTCAGCAAT | |
| MdVIT9 | ACCGCGAGAAGCAAGCACT | AGCGAACGGCACGGTAAGA | |
| 克隆基因及 启动子 Clone primers | MD04G1208900 | ATGGCTTCCCAACGTGACCA | TCAGATTCCCAAACCACTAGACC |
| MD05G1023100 | ATGTGTCCTCCACACTTCTCCAA | TCATAGCCCACTTGAGCCAATC | |
| MD12G1223300 | ATGGCTTCCCAACGTGACCA | TCACACCTGCATACCACTAGAC |
Table 1 qRT-PCR and clone primers for expression on analysis of VIT in apple
| 用途Usage | 基因Gene | 上游引物(5′-3′)Forward primer | 下游引物(5′-3′)Reverse primer |
|---|---|---|---|
| qRT-PCR | MdVIT1 | CTCCAATCCCCGAAGACGA | GCGATGCAACGGAAACCA |
| MdVIT2 | TCCGCTTCTCCCGTTAATATT | CGAGGTCGTTTGTGGTCATT | |
| MdVIT3 | CACCAAACAACGCTAGACCAA | TCACAGCTCCGACTCCCAT | |
| MdVIT4 | TTCGATTACTCCCAGAGGACAC | AGCGATGCAACAGAAACCAG | |
| MdVIT5 | AATCCCAGAAGGCGACGAA | GAGCGATGCAACAGAAACCAGCCCA | |
| MdVIT6 | TCTTACCGTGCCGTTTGCT | ATATCCGCCGAGTCCCATG | |
| MdVIT7 | CAAGGACCGAGCAAGTGCA | CATCAAGGACGTGGTTGACAGT | |
| MdVIT8 | CGACGGTCTCACTGTCCCTT | GGCGGCAACTTCAGCAAT | |
| MdVIT9 | ACCGCGAGAAGCAAGCACT | AGCGAACGGCACGGTAAGA | |
| 克隆基因及 启动子 Clone primers | MD04G1208900 | ATGGCTTCCCAACGTGACCA | TCAGATTCCCAAACCACTAGACC |
| MD05G1023100 | ATGTGTCCTCCACACTTCTCCAA | TCATAGCCCACTTGAGCCAATC | |
| MD12G1223300 | ATGGCTTCCCAACGTGACCA | TCACACCTGCATACCACTAGAC |
| 基因名 Gene name | 登录号 Gene ID | 序列位置 Sequence position | 染色体 位置 Chr | 开放阅 读区/bp ORF | 氨基酸 数/aa Length of amino acid | 分子量/ kD Molecular weigh | 不稳定 系数 Instability index | 理论等 电点 pI | 蛋白质 疏水性 Hydroph- obicity | 脂肪指数 Aliphatic index | 跨膜结 构域 TMD |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MdVIT1 | MD04G1208900 | 29415642..29416325 | Chr04 | 684 | 227 | 23.71 | 42.66 | 6.13 | 0.29 | 97.14 | 4 |
| MdVIT2 | MD05G1023100 | 3872852..3873643 | Chr05 | 792 | 263 | 27.68 | 41.63 | 6.96 | 0.25 | 97.91 | 5 |
| MdVIT3 | MD10G1024000 | 3007772..3008458 | Chr10 | 687 | 228 | 24.01 | 34.11 | 6.13 | 0.28 | 101.05 | 4 |
| MdVIT4 | MD12G1223300 | 30004855..30005538 | Chr12 | 684 | 227 | 23.87 | 43.44 | 7.78 | 0.30 | 96.26 | 4 |
| MdVIT5 | MD12G1223400 | 30015123..30015806 | Chr12 | 684 | 227 | 23.90 | 45.02 | 6.30 | 0.28 | 96.26 | 4 |
| MdVIT6 | MD13G1018700 | 1174829..1178506 | Chr13 | 744 | 247 | 26.36 | 36.99 | 5.18 | 0.19 | 101.30 | 4 |
| MdVIT7 | MD15G1227000 | 18449646..18451051 | Chr15 | 798 | 265 | 27.99 | 42.77 | 8.34 | 0.01 | 89.51 | 5 |
| MdVIT8 | MD16G1016500 | 1240143..1242202 | Chr16 | 747 | 248 | 26.52 | 36.31 | 5.00 | 0.38 | 105.16 | 4 |
| MdVIT9 | MD16G1016600 | 1243044..1245822 | Chr16 | 744 | 247 | 26.41 | 35.57 | 5.89 | 0.21 | 105.63 | 4 |
Table 2 Detailed information of identified VIT genes in apple genome
| 基因名 Gene name | 登录号 Gene ID | 序列位置 Sequence position | 染色体 位置 Chr | 开放阅 读区/bp ORF | 氨基酸 数/aa Length of amino acid | 分子量/ kD Molecular weigh | 不稳定 系数 Instability index | 理论等 电点 pI | 蛋白质 疏水性 Hydroph- obicity | 脂肪指数 Aliphatic index | 跨膜结 构域 TMD |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MdVIT1 | MD04G1208900 | 29415642..29416325 | Chr04 | 684 | 227 | 23.71 | 42.66 | 6.13 | 0.29 | 97.14 | 4 |
| MdVIT2 | MD05G1023100 | 3872852..3873643 | Chr05 | 792 | 263 | 27.68 | 41.63 | 6.96 | 0.25 | 97.91 | 5 |
| MdVIT3 | MD10G1024000 | 3007772..3008458 | Chr10 | 687 | 228 | 24.01 | 34.11 | 6.13 | 0.28 | 101.05 | 4 |
| MdVIT4 | MD12G1223300 | 30004855..30005538 | Chr12 | 684 | 227 | 23.87 | 43.44 | 7.78 | 0.30 | 96.26 | 4 |
| MdVIT5 | MD12G1223400 | 30015123..30015806 | Chr12 | 684 | 227 | 23.90 | 45.02 | 6.30 | 0.28 | 96.26 | 4 |
| MdVIT6 | MD13G1018700 | 1174829..1178506 | Chr13 | 744 | 247 | 26.36 | 36.99 | 5.18 | 0.19 | 101.30 | 4 |
| MdVIT7 | MD15G1227000 | 18449646..18451051 | Chr15 | 798 | 265 | 27.99 | 42.77 | 8.34 | 0.01 | 89.51 | 5 |
| MdVIT8 | MD16G1016500 | 1240143..1242202 | Chr16 | 747 | 248 | 26.52 | 36.31 | 5.00 | 0.38 | 105.16 | 4 |
| MdVIT9 | MD16G1016600 | 1243044..1245822 | Chr16 | 744 | 247 | 26.41 | 35.57 | 5.89 | 0.21 | 105.63 | 4 |
| 蛋白 | 液泡 | 质膜 | 内质网 | 高尔基体 | 胞外 |
|---|---|---|---|---|---|
| Protein | Vacuolar | Plasma membrane | Endoplasmic reticulum | Golgi apparatus | Extracellular |
| MdVIT1 | 9 | 4 | 1 | ||
| MdVIT2 | 2 | 7 | 1 | ||
| MdVIT3 | 12 | 2 | |||
| MdVIT4 | 4 | 5.5 | 1 | 2 | |
| MdVIT5 | 7 | 5 | 1 | 1 | |
| MdVIT6 | 1 | 12 | |||
| MdVIT7 | 2 | 5 | 6 | 1 | |
| MdVIT8 | 2 | 8 | 3 | 1 | |
| MdVIT9 | 2 | 12 |
Table 3 Subcellular predication of apple MdVIT
| 蛋白 | 液泡 | 质膜 | 内质网 | 高尔基体 | 胞外 |
|---|---|---|---|---|---|
| Protein | Vacuolar | Plasma membrane | Endoplasmic reticulum | Golgi apparatus | Extracellular |
| MdVIT1 | 9 | 4 | 1 | ||
| MdVIT2 | 2 | 7 | 1 | ||
| MdVIT3 | 12 | 2 | |||
| MdVIT4 | 4 | 5.5 | 1 | 2 | |
| MdVIT5 | 7 | 5 | 1 | 1 | |
| MdVIT6 | 1 | 12 | |||
| MdVIT7 | 2 | 5 | 6 | 1 | |
| MdVIT8 | 2 | 8 | 3 | 1 | |
| MdVIT9 | 2 | 12 |
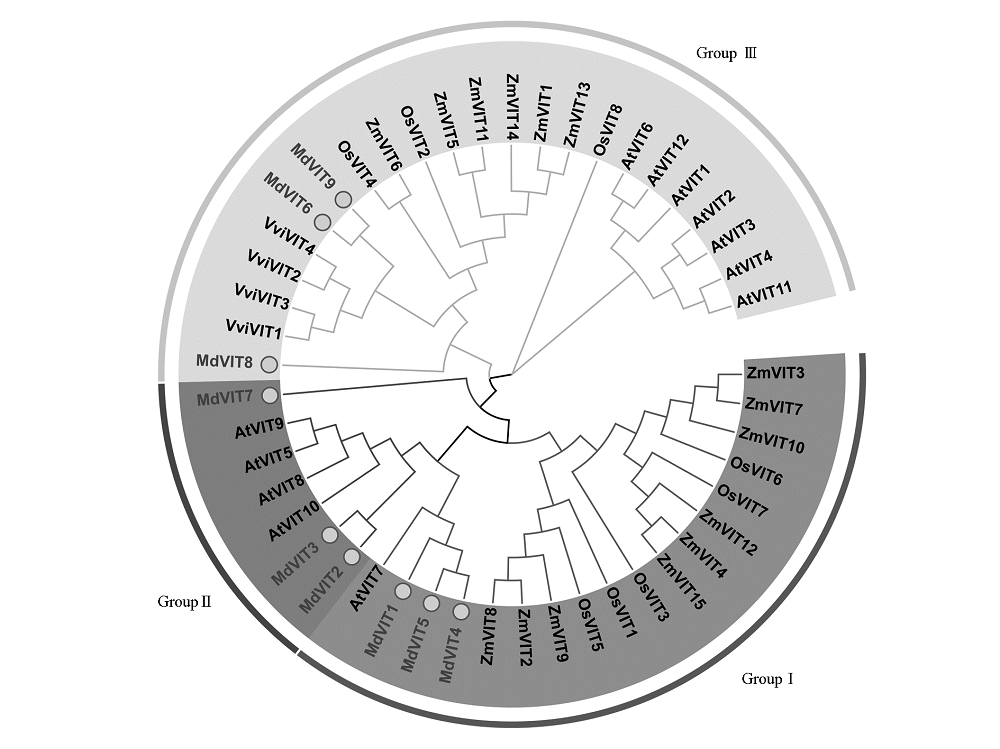
Fig. 2 The phylogenetic tree of VIT gene family in Arabidopsis thaliana(At),Oryza sativa(Os),Zea mays(Zm), Vitis vinifera(Vv)and Malus × domestica(Md)
| 基因 Gene | 基因 Gene | 同义 替换率 Ka | 非同义 替换率 Ks | 选择性 压力 Ka/Ks | 分离时间/Mya Divergence Time | 复制类型 Duplication Types |
|---|---|---|---|---|---|---|
| MdVIT1(MD04G1208900) | MdVIT5(MD12G1223400) | 0.055 | 0.226 | 0.245 | 7.527 | 片段复制 Segmental duplication |
| MdVIT1(MD04G1208900) | MdVIT4(MD12G1223300) | 0.071 | 0.190 | 0.371 | 6.347 | 片段复制 Segmental duplication |
| MdVIT2(MD05G1023100) | MdVIT3(MD10G1024000) | 1.030 | 0.912 | 1.130 | 30.387 | 串联重复 Tandem dupication |
| MdVIT4(MD12G1223300) | MdVIT5(MD12G1223400) | 0.036 | 0.103 | 0.349 | 3.425 | 片段复制 Segmental duplication |
Table 4 Divergence time of the VIT paralogues in apple
| 基因 Gene | 基因 Gene | 同义 替换率 Ka | 非同义 替换率 Ks | 选择性 压力 Ka/Ks | 分离时间/Mya Divergence Time | 复制类型 Duplication Types |
|---|---|---|---|---|---|---|
| MdVIT1(MD04G1208900) | MdVIT5(MD12G1223400) | 0.055 | 0.226 | 0.245 | 7.527 | 片段复制 Segmental duplication |
| MdVIT1(MD04G1208900) | MdVIT4(MD12G1223300) | 0.071 | 0.190 | 0.371 | 6.347 | 片段复制 Segmental duplication |
| MdVIT2(MD05G1023100) | MdVIT3(MD10G1024000) | 1.030 | 0.912 | 1.130 | 30.387 | 串联重复 Tandem dupication |
| MdVIT4(MD12G1223300) | MdVIT5(MD12G1223400) | 0.036 | 0.103 | 0.349 | 3.425 | 片段复制 Segmental duplication |
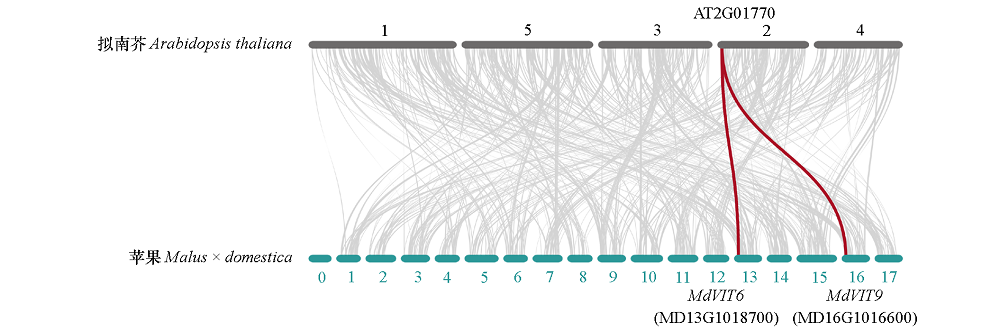
Fig. 5 Synteny analysis of VIT genes between apple and Arabidopsis thaliana Gray lines in the background indicate the collinear blocks with apple and Arabidopsis thaliana,while the red lines highlight the syntenic VIT gene pairs.

Fig. 7 Expression profiles of MdVIT genes in different tissues and different apple varieties GD reference‘Golden Delicious’;M69,M74,M20,X8877,M14,M49,X4102,X4442 × X2596,and X3069 × X992 were various apple hybrids.
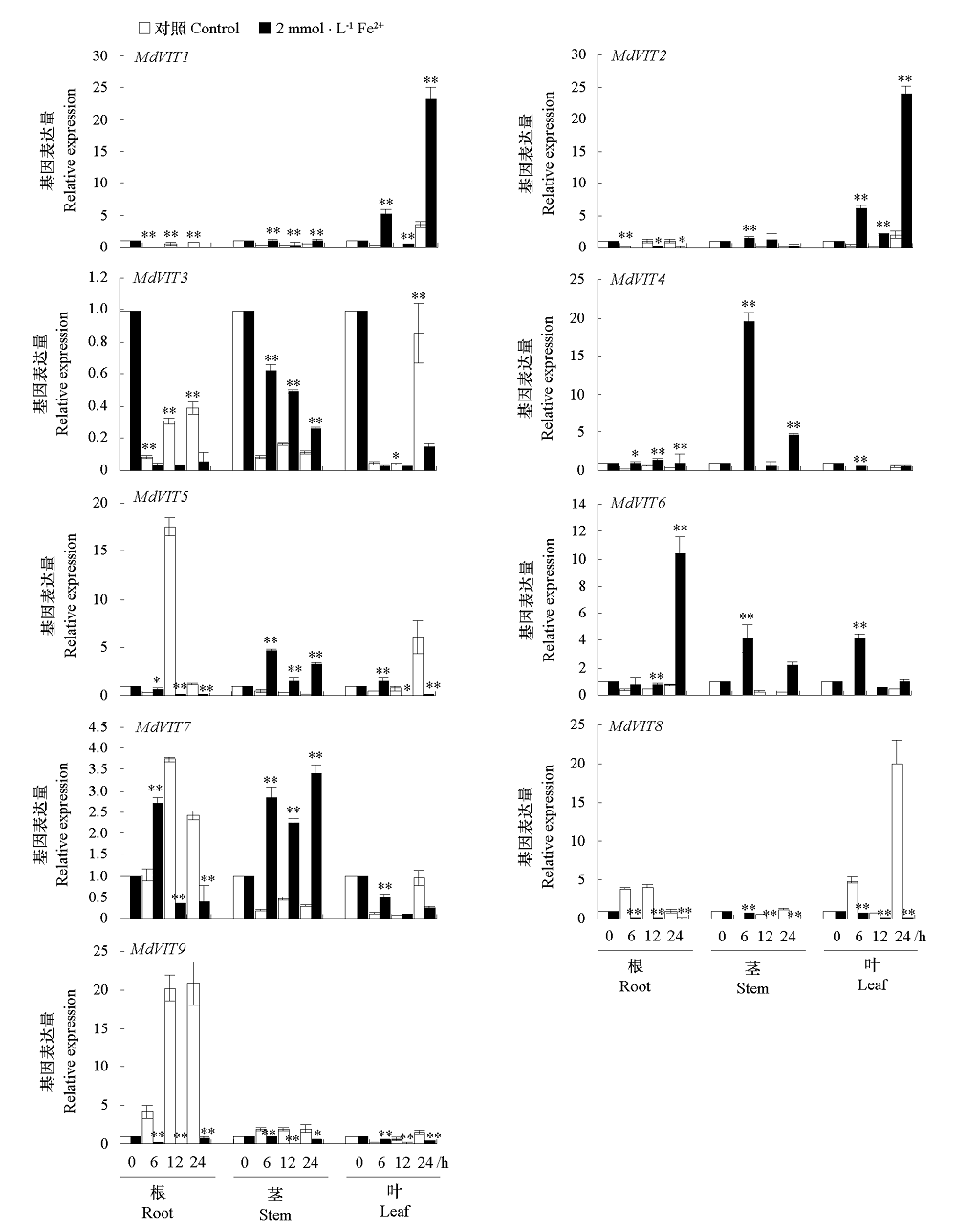
Fig. 8 Expression profile of MdVIT in different tissues in apple Significance difference between treatment and the control were analyzed by t-test. * α = 0.05,** α = 0.01.
| [1] |
Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, Duvaud S, Flegel V, Fortier A, Gasteiger E. 2012. ExPASy:SIB bioinformatics resource portal. Nucleic Acids Research, 40(W1):W597-W603.
doi: 10.1093/nar/gks400 URL |
| [2] |
Bailey T L, Boden M, Buske F A, Frith M, Grant C E, Clementi L, Ren J Y, Li W W, Noble W S. 2009. MEME SUITE:tools for motif discovery and searching. Nucleic Acids Research, 37:W202-W208.
doi: 10.1093/nar/gkp335 URL |
| [3] |
Blanc G, Wolfe K H. 2004. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. The Plant Cell, 16(7):1667-1678.
doi: 10.1105/tpc.021345 URL |
| [4] |
Cao J, Li X. 2015. Identification and phylogenetic analysis of late embryogenesis abundant proteins family in tomato(Solanum lycopersicum). Planta, 241(3):757-772.
doi: 10.1007/s00425-014-2215-y URL |
| [5] |
Cao J. 2019. Molecular evolution of the vacuolar iron transporter(VIT)family genes in 14 plant species. Genes, 10(2):16.
doi: 10.3390/genes10010016 URL |
| [6] |
Colmenero-Flores J M, Martinez G, Gamba G, Vazquez N, Iglesias D J, Brumos J, Talon M. 2007. Identification and functional characterization of cation-chloride cotransporters in plants. Plant Journal, 50(2):278-292.
pmid: 17355435 |
| [7] | Connolly E L, Guerinot M L. 2002. Iron stress in plants. Genome Biology, 3(8):1-4. |
| [8] |
Connorton J M, Jones E R, Rodriguez-Ramiro I, Fairweather-Tait S, Uauy C, Balk J. 2017. Wheat vacuolar iron transporter Ta VIT2 transports Fe and Mn and is effective for biofortification. Plant Physiology, 174(4):2434-2444.
doi: 10.1104/pp.17.00672 pmid: 28684433 |
| [9] |
Crombach A, Hogeweg P. 2007. Chromosome rearrangements and the evolution of genome structuring and adaptability. Molecular Biology and Evolution, 24(5):1130-1139.
doi: 10.1093/molbev/msm033 URL |
| [10] | Grotz N, Guerinot M L. 2006. Molecular aspects of Cu,Fe and Zn homeostasis in plants. Biochimica Et Biophysica Acta- Molecular Cell Research, 1763(7):595-608. |
| [11] |
Gu Z L, Cavalcanti A, Chen F C, Bouman P, Li W H. 2002. Extent of gene duplication in the genomes of Drosophila,nematode,and yeast. Molecular Biology and Evolution, 19(3):256-262.
doi: 10.1093/oxfordjournals.molbev.a004079 URL |
| [12] |
Hell R, Stephan U W. 2003. Iron uptake,trafficking and homeostasis in plants. Planta, 216(4):541-551.
doi: 10.1007/s00425-002-0920-4 URL |
| [13] |
Hu B, Jin J P, Guo A Y, Zhang H, Luo J C, Gao G. 2015. GSDS 2.0:an upgraded gene feature visualization server. Bioinformatics, 31(8):1296-1297.
doi: 10.1093/bioinformatics/btu817 URL |
| [14] |
Imsande J. 1998. Iron,sulfur,and chlorophyll deficiencies:a need for an integrative approach in plant physiology. Physiologia Plantarum, 103(1):139-144.
doi: 10.1034/j.1399-3054.1998.1030117.x URL |
| [15] |
Kim S A, Punshon T, Lanzirotti A, Li L T, Alonso J M, Ecker J R, Kaplan J, Guerinot M L. 2006. Localization of iron in Arabidopsis seed requires the vacuolar membrane transporter VIT1. Science, 314(5803):1295-1298.
doi: 10.1126/science.1132563 URL |
| [16] |
Kong X Q, Gao X H, Sun W, An J, Zhao Y X, Zhang H. 2011. Cloning and functional characterization of a cation-chloride cotransporter gene OsCCC1. Plant Molecular Biology, 75(6):567-578.
doi: 10.1007/s11103-011-9744-6 URL |
| [17] |
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones S J, Marra M A. 2009. Circos: An information aesthetic for comparative genomics. Genome Res, 19(9):1639-1645.
doi: 10.1101/gr.092759.109 URL |
| [18] | Latrasse D, Rodriguez-Granados N Y, Veluchamy A, Mariappan K G, Bevilacqua C, Crapart N, Camps C, Sommard V, Raynaud C, Dogimont C. 2017. The quest for epigenetic regulation underlying unisexual flower development in Cucumis melo. Epigenetics & Chromatin, 10. |
| [19] |
Le Hir H, Nott A, Moore M J. 2003. How introns influence and enhance eukaryotic gene expression. Trends Biochem Sci, 28(4):215-220.
doi: 10.1016/S0968-0004(03)00052-5 URL |
| [20] |
Lee T H, Tang H B, Wang X Y, Paterson A H. 2013. PGDD:a database of gene and genome duplication in plants. Nucleic Acids Research, 41(D1):D1152-D1158.
doi: 10.1093/nar/gks1104 URL |
| [21] |
Li L T, Chen O S, Ward DM, Kaplan J. 2001. CCC1 is a transporter that mediates vacuolar iron storage in yeast. J Biol Chem, 276(31):29515-29519.
pmid: 11390404 |
| [22] | Li S, Wang N, Ji D D, Xue Z Y, Yu Y C, Jiang Y P, Liu J L, Liu Z H, Xiang F N. 2016. Evolutionary and functional analysis of membrane-bound NAC transcription factor genes in soybean. Plant Physiology, 172(3):1804-1820. |
| [23] |
Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆CT method. Methods, 25(4):402-408.
pmid: 11846609 |
| [24] |
Mattick J S, Gagen M J. 2001. The evolution of controlled multitasked gene networks: The role of introns and other noncoding RNAs in the development of complex organisms. Molecular Biology and Evolution, 18(9):1611-1630.
doi: 10.1093/oxfordjournals.molbev.a003951 URL |
| [25] |
Mistry J, Finn R D, Eddy S R, Bateman A, Punta M. 2013. Challenges in homology search:HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Research, 41(12):e121-e121.
doi: 10.1093/nar/gkt263 URL |
| [26] |
Moes A D, van der Lubbe N, Zietse R, Loffing J, Hoorn E J. 2014. The sodium chloride cotransporter SLC12A3: new roles in sodium,potassium,and blood pressure regulation. Pflugers Archiv-European Journal of Physiology, 466(1):107-118.
doi: 10.1007/s00424-013-1407-9 URL |
| [27] |
Nagasaka S, Takahashi M, Nakanishi-Itai R, Bashir K, Nakanishi H, Mori S, Nishizawa N K. 2009. Time course analysis of gene expression over 24 h in Fe-deficient barley roots. Plant Mol Biol, 69(5):621-631.
doi: 10.1007/s11103-008-9443-0 pmid: 19089316 |
| [28] | Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution, 4(4):406-425. |
| [29] |
Schmidt W. 2003. Iron solutions: acquisition strategies and signaling pathways in plants. Trends in Plant Science, 8(4):188-193.
doi: 10.1016/S1360-1385(03)00048-7 URL |
| [30] |
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5:molecular evolutionary genetics analysis using maximum likelihood,evolutionary distance,and maximum parsimony methods. Molecular Biology and Evolution, 28(10):2731-2739.
doi: 10.1093/molbev/msr121 URL |
| [31] | Wang Ruonan, Nie Lanchun, Zhang Shuangshuang, Cui Qiang, Jia Mingfei. 2019. Research progress on plant resistance to heavy metal stress. Acta Horticulturae Sinica, 46(1):157-170. (in Chinese) |
| 王若男, 乜兰春, 张双双, 崔强, 贾明飞. 2019. 植物抗重金属胁迫研究进展. 园艺学报, 46(1):157-170. | |
| [32] | Wang Y P, Tang H B, DeBarry J D, Tan X, Li J P, Wang X Y, Lee T H, Jin H Z, Marler B, Guo H. 2012. MCScanX:a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Research, 40(7):14. |
| [33] | Xu Rui-rui, Li Rui, Wang Xiao-fei, Hao Yu-jin, 2018. Identification and expression analysis under abiotic stresses of OFP gene family in apple. Scientia Agricultura Sinica, 51(10):141-152. (in Chinese) |
| 许瑞瑞, 李睿, 王小非, 郝玉金. 2018. 苹果OFP基因家族的全基因组鉴定与非生物逆境表达分析. 中国农业科学, 51(10):141-152. | |
| [34] |
Yang ZF, Zhou Y, Wang X F, Gu S L, Yu J M, Liang G H, Yan C J, Xu C W. 2008. Genomewide comparative phylogenetic and molecular evolutionary analysis of tubby-like protein family in Arabidopsis,rice,and poplar. Genomics, 92(4):246-253.
doi: 10.1016/j.ygeno.2008.06.001 URL |
| [35] |
Zhang Y, Xu Y H, Yi H Y, Gong J M. 2012. Vacuolar membrane transporters OsVIT1 and OsVIT2 modulate iron translocation between flag leaves and seeds in rice. Plant Journal, 72(3):400-410.
doi: 10.1111/tpj.2012.72.issue-3 URL |
| [36] | Zhou Rong-fang, Yuan Wu-zhou, Tong Chao-bo, Huang Jun-yan, Cheng Xiao-Hui, Yu Jing-yin, Dong Cai-hua, Liu Yue-ying, Liu Sheng-yi. 2014. Identification and evolution of VIT gene family between A and C genomes in Brassica. Chinese Journal of Oil Crop Sciences, 36(5):551-561. (in Chinese) |
| 周荣芳, 袁午舟, 童超波, 黄军艳, 程晓晖, 于景印, 董彩华, 刘越英, 刘胜毅. 2014. 芸薹属AC基因组中VIT基因家族的鉴定与进化. 中国油料作物学报, 36(5):551-561. |
| [1] | WANG Xiaochen, NIE Ziye, LIU Xianju, DUAN Wei, FAN Peige, and LIANG Zhenchang, . Effects of Abscisic Acid on Monoterpene Synthesis in‘Jingxiangyu’Grape Berries [J]. Acta Horticulturae Sinica, 2023, 50(2): 237-249. |
| [2] | YU Tingting, LI Huan, NING Yuansheng, SONG Jianfei, PENG Lulin, JIA Junqi, ZHANG Weiwei, and YANG Hongqiang. Genome-wide Identification of GRAS Gene Family in Apple and Expression Analysis of Its Response to Auxin [J]. Acta Horticulturae Sinica, 2023, 50(2): 397-409. |
| [3] | ZHAI Hanhan, ZHAI Yujie, TIAN Yi, ZHANG Ye, YANG Li, WEN Zhiliang, CHEN Haijiang. Genome-wide Identification of Peach SAUR Gene Family and Characterization of PpSAUR5 Gene [J]. Acta Horticulturae Sinica, 2023, 50(1): 1-14. |
| [4] | HAN Xiaolei, ZHANG Caixia, LIU Kai, YANG An, YAN Jiadi, LI Wuxing, KANG Liqun, and CONG Peihua. A New Mid-ripening Apple Cultivar‘Zhongping Youlei’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 1-2. |
| [5] | SUN Simiao, WANG Kun, GAO Yuan, WANG Dajiang, and LI Lianwen. A New Ornamental Crabapple Cultivar‘Zichen’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 267-268. |
| [6] | HAN Xiaolei, ZHANG Caixia, LIU Kai, YAN Jiadi, LI Wuxing, KANG Liqun, and CONG Peihua. A New Mid-ripening Apple Cultivar‘Pingyou 2’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 1-2. |
| [7] | WANG Qiang, CONG Peihua, and LIU Xiaofeng. A New Late Ripening Apple Cultivar‘Huayou Tianwa’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 3-4. |
| [8] | WANG Qiang, CONG Peihua, and LIU Xiaofeng. A New Mid-ripening Apple Cultivar‘Huayou Baomi’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 5-6. |
| [9] | YANG Ling, CONG Peihua, WANG Qiang, LI Wuxing, and KANG Liqun. A New Mid-ripening Apple Cultivar‘Huafeng’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 7-8. |
| [10] | LIU Chuanhe, HE Han, SHAO Xuehua, LAI Duo, KUANG Shizi, XIAO Weiqiang, LIU Yan. A New Pineapple Cultivar‘Yuetong’ [J]. Acta Horticulturae Sinica, 2022, 49(9): 2053-2054. |
| [11] | GAO Yanlong, WU Yuxia, ZHANG Zhongxing, WANG Shuangcheng, ZHANG Rui, ZHANG De, WANG Yanxiu. Bioinformatics Analysis of Apple ELO Gene Family and Its Expression Analysis Under Low Temperature Stress [J]. Acta Horticulturae Sinica, 2022, 49(8): 1621-1636. |
| [12] | LIU Chaoyang, LIAO Zhichan, LU Xinxin, HE Yehua. Identification of CslD Gene Family in Pineapple and Functional Analysis of AcoCslD2a [J]. Acta Horticulturae Sinica, 2022, 49(8): 1650-1662. |
| [13] | ZHENG Xiaodong, XI Xiangli, LI Yuqi, SUN Zhijuan, MA Changqing, HAN Mingsan, LI Shaoxuan, TIAN Yike, WANG Caihong. Effects and Regulating Mechanism of Exogenous Brassinosteroids on the Growth of Malus hupehensis Under Saline-alkali Stress [J]. Acta Horticulturae Sinica, 2022, 49(7): 1401-1414. |
| [14] | XIA Yan, HUANG Song, WU Xueli, LIU Yiqi, WANG Miaomiao, SONG Chunhui, BAI Tuanhui, SONG Shangwei, PANG Hongguang, JIAO Jian, ZHENG Xianbo. Identification and Analysis of Apple Viruses Diseases Based on Virome Sequencing Technology [J]. Acta Horticulturae Sinica, 2022, 49(7): 1415-1428. |
| [15] | LIU Zhaoxia, ZHANG Xin, WANG Lu, MA Yuting, CHEN Qian, ZHU Zhanling, GE Shunfeng, JIANG Yuanmao. Effects of Fertilizer Hole Application Sites on Fine Root Growth,15N Absorption and Utilization,Yield and Quality of Apple Trees [J]. Acta Horticulturae Sinica, 2022, 49(7): 1545-1556. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd