
Acta Horticulturae Sinica ›› 2021, Vol. 48 ›› Issue (1): 83-95.doi: 10.16420/j.issn.0513-353x.2020-0042
• Research Papers • Previous Articles Next Articles
JIAN Hongju1, WAN Mengyuan1, FU Yi1, DING Yi1, JU Xisan1, SHANG Lina1, LI Hui1, WANG Jichun1, HU Bogeng2, LÜ Dianqiu1,**( )
)
Received:2020-03-30
Revised:2020-06-02
Online:2021-01-25
Published:2021-01-29
Contact:
Lü Dianqiu
E-mail:smallpotatoes@126.com
CLC Number:
JIAN Hongju, WAN Mengyuan, FU Yi, DING Yi, JU Xisan, SHANG Lina, LI Hui, WANG Jichun, HU Bogeng, LÜ Dianqiu. Genome-wide Identification and Expression Analysis of Chitinase Gene Family in Potato and Its Response to Exogenous Salicylic Acid and Jasmonic Acid[J]. Acta Horticulturae Sinica, 2021, 48(1): 83-95.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2020-0042
| 基因名称 Gene | 引物序列(5′-3′) Primer sequence |
|---|---|
| StuEFLa | F:ATTGGAAACGGATATGCTCCA;R:TCCTTACCTGAACGCCTGTCA |
| StuChi7 | F:CGAATACATTGAACAAAACGCG;R:GTATCATTCTTCGTTGGCTTCC |
| StuChi11 | F:CGTCATCTCAAATTCCGTGTTT;R:CAGTAGTGTTTTGGGAGGTTTG |
| StuChi17 | F:CTAAACGACTTCCTGGTTTTGG;R:CATTGAGTCAACATCACCTTGG |
| StuChi21 | F:ACTTCCCATGAAACTACTGGAG;R:TGCTCTCTAAGGAAGCAGTAAC |
Table 1 Primer sequences
| 基因名称 Gene | 引物序列(5′-3′) Primer sequence |
|---|---|
| StuEFLa | F:ATTGGAAACGGATATGCTCCA;R:TCCTTACCTGAACGCCTGTCA |
| StuChi7 | F:CGAATACATTGAACAAAACGCG;R:GTATCATTCTTCGTTGGCTTCC |
| StuChi11 | F:CGTCATCTCAAATTCCGTGTTT;R:CAGTAGTGTTTTGGGAGGTTTG |
| StuChi17 | F:CTAAACGACTTCCTGGTTTTGG;R:CATTGAGTCAACATCACCTTGG |
| StuChi21 | F:ACTTCCCATGAAACTACTGGAG;R:TGCTCTCTAAGGAAGCAGTAAC |
| 基因 Gene | 基因编号 Gene ID | 位置 Location | 氨基酸数 Number of amino acid | 分子量/D MW | pI | 外显子数Exon No. | 亚细胞定位Subcellular location | 分类 Subgroup | 亚家族Subfamily |
|---|---|---|---|---|---|---|---|---|---|
| StuChi1 | PGSC0003DMG400000073 | chr01: 72805788-72807273 | 298 | 31 990.93 | 4.5 | 1 | 细胞外 Outside | C | GH18 |
| StuChi2 | PGSC0003DMG400000074 | chr01: 72808827-72810061 | 298 | 31 821.89 | 4.6 | 1 | 细胞外 Outside | C | GH18 |
| StuChi3 | PGSC0003DMG400000078 | chr01: 72833577-72842485 | 302 | 32 363.59 | 5.9 | 1 | 细胞外 Outside | C | GH18 |
| StuChi4 | PGSC0003DMG400001528 | chr02: 38744461-38745679 | 252 | 27 535.83 | 5.9 | 3 | 细胞外 Outside | F | GH19 |
| StuChi5 | PGSC0003DMG400001529 | chr02: 38749197-38750328 | 254 | 27 389.43 | 4.7 | 3 | 细胞外 Outside | F | GH19 |
| StuChi6 | PGSC0003DMG400003191 | chr07: 334428-335534 | 368 | 41 516.91 | 7.1 | 1 | 细胞外 Outside | A | GH18 |
| StuChi7 | PGSC0003DMG400004593 | chr12: 59618049-59621290 | 327 | 36 260.51 | 7.0 | 3 | 细胞外 Outside | D | GH19 |
| StuChi8 | PGSC0003DMG400006591 | chr03: 44670486-44671710 | 294 | 32 248.04 | 9.3 | 1 | 质膜Plasma membrane | C | GH18 |
| StuChi9 | PGSC0003DMG400008673 | chr02: 19537307-19539201 | 264 | 28 748.54 | 8.8 | 3 | 细胞外 Outside | F | GH19 |
| 基因 Gene | 基因编号 Gene ID | 位置 Location | 氨基酸数 Number of amino acid | 分子量/D MW | pI | 外显子数Exon No. | 亚细胞定位Subcellular location | 分类 Subgroup | 亚家族Subfamily |
| StuChi10 | PGSC0003DMG400008796 | chr00: 25050182-25052792 | 323 | 35 031.61 | 6.5 | 3 | 细胞外 Outside | F | GH19 |
| StuChi11 | PGSC0003DMG400008797 | chr00: 25056603-25057931 | 280 | 29 350.85 | 8.5 | 4 | 细胞外 Outside | F | GH19 |
| StuChi12 | PGSC0003DMG400011836 | chr07: 292990-294141 | 383 | 41 627.01 | 4.8 | 1 | 过氧化物酶体 Microbody (Peroxisome) | B | GH18 |
| StuChi13 | PGSC0003DMG400011842 | chr07: 294672-296216 | 355 | 39 811.97 | 9.2 | 2 | 过氧化物酶体 Microbody (Peroxisome) | B | GH18 |
| StuChi14 | PGSC0003DMG400011843 | chr07: 303868-306084 | 372 | 42 305.99 | 9.3 | 3 | 线粒体基质 Mitochondrial matrix space | A | GH18 |
| StuChi15 | PGSC0003DMG400016302 | chr06: 39116330-39117653 | 285 | 31 468.23 | 5.9 | 2 | 细胞外 Outside | E | GH19 |
| StuChi16 | PGSC0003DMG400016302 | chr06: 39116330-39117653 | 136 | 15 182.02 | 5.4 | 1 | 细胞质 Cytoplasm | E | GH19 |
| StuChi17 | PGSC0003DMG400021602 | chr09: 61360398-61361921 | 323 | 35 986.30 | 7.9 | 3 | 细胞外 Outside | D | GH19 |
| StuChi18 | PGSC0003DMG400025063 | chr04: 61242553-61244259 | 276 | 29 982.95 | 4.6 | 2 | 细胞外 Outside | E | GH19 |
| StuChi19 | PGSC0003DMG400025063 | chr04: 61242553-61244259 | 136 | 14 988.53 | 4.3 | 1 | 细胞质 Cytoplasm | E | GH19 |
| StuChi20 | PGSC0003DMG400026853 | chr00: 21407639-21408792 | 316 | 34 091.35 | 5.2 | 3 | 质膜Plasma membrane | F | GH19 |
| StuChi21 | PGSC0003DMG400026854 | chr00: 21415307-21416600 | 320 | 34 117.04 | 6.2 | 3 | 细胞外 Outside | F | GH19 |
| StuChi22 | PGSC0003DMG400026855 | chr00: 21419777-21420932 | 332 | 35 506.90 | 8.4 | 3 | 细胞外 Outside | F | GH19 |
| StuChi23 | PGSC0003DMG400033882 | chr05: 44304719-44306659 | 292 | 30 945.16 | 7.5 | 2 | 细胞外 Outside | C | GH18 |
| StuChi24 | PGSC0003DMG400037480 | chr07: 347182-349354 | 435 | 49 224.80 | 6.7 | 4 | 质膜Plasma membrane | A | GH18 |
| StuChi25 | PGSC0003DMG400040317 | chr00: 21299520-21300925 | 279 | 29 606.22 | 8.4 | 2 | 细胞外 Outside | F | GH19 |
| StuChi26 | PGSC0003DMG402001531 | chr02: 38760083-38761491 | 263 | 28 819.75 | 9.2 | 3 | 细胞外 Outside | F | GH19 |
Table 2 Genome-wide identification and molecular characterization of chitinase gene family in potato
| 基因 Gene | 基因编号 Gene ID | 位置 Location | 氨基酸数 Number of amino acid | 分子量/D MW | pI | 外显子数Exon No. | 亚细胞定位Subcellular location | 分类 Subgroup | 亚家族Subfamily |
|---|---|---|---|---|---|---|---|---|---|
| StuChi1 | PGSC0003DMG400000073 | chr01: 72805788-72807273 | 298 | 31 990.93 | 4.5 | 1 | 细胞外 Outside | C | GH18 |
| StuChi2 | PGSC0003DMG400000074 | chr01: 72808827-72810061 | 298 | 31 821.89 | 4.6 | 1 | 细胞外 Outside | C | GH18 |
| StuChi3 | PGSC0003DMG400000078 | chr01: 72833577-72842485 | 302 | 32 363.59 | 5.9 | 1 | 细胞外 Outside | C | GH18 |
| StuChi4 | PGSC0003DMG400001528 | chr02: 38744461-38745679 | 252 | 27 535.83 | 5.9 | 3 | 细胞外 Outside | F | GH19 |
| StuChi5 | PGSC0003DMG400001529 | chr02: 38749197-38750328 | 254 | 27 389.43 | 4.7 | 3 | 细胞外 Outside | F | GH19 |
| StuChi6 | PGSC0003DMG400003191 | chr07: 334428-335534 | 368 | 41 516.91 | 7.1 | 1 | 细胞外 Outside | A | GH18 |
| StuChi7 | PGSC0003DMG400004593 | chr12: 59618049-59621290 | 327 | 36 260.51 | 7.0 | 3 | 细胞外 Outside | D | GH19 |
| StuChi8 | PGSC0003DMG400006591 | chr03: 44670486-44671710 | 294 | 32 248.04 | 9.3 | 1 | 质膜Plasma membrane | C | GH18 |
| StuChi9 | PGSC0003DMG400008673 | chr02: 19537307-19539201 | 264 | 28 748.54 | 8.8 | 3 | 细胞外 Outside | F | GH19 |
| 基因 Gene | 基因编号 Gene ID | 位置 Location | 氨基酸数 Number of amino acid | 分子量/D MW | pI | 外显子数Exon No. | 亚细胞定位Subcellular location | 分类 Subgroup | 亚家族Subfamily |
| StuChi10 | PGSC0003DMG400008796 | chr00: 25050182-25052792 | 323 | 35 031.61 | 6.5 | 3 | 细胞外 Outside | F | GH19 |
| StuChi11 | PGSC0003DMG400008797 | chr00: 25056603-25057931 | 280 | 29 350.85 | 8.5 | 4 | 细胞外 Outside | F | GH19 |
| StuChi12 | PGSC0003DMG400011836 | chr07: 292990-294141 | 383 | 41 627.01 | 4.8 | 1 | 过氧化物酶体 Microbody (Peroxisome) | B | GH18 |
| StuChi13 | PGSC0003DMG400011842 | chr07: 294672-296216 | 355 | 39 811.97 | 9.2 | 2 | 过氧化物酶体 Microbody (Peroxisome) | B | GH18 |
| StuChi14 | PGSC0003DMG400011843 | chr07: 303868-306084 | 372 | 42 305.99 | 9.3 | 3 | 线粒体基质 Mitochondrial matrix space | A | GH18 |
| StuChi15 | PGSC0003DMG400016302 | chr06: 39116330-39117653 | 285 | 31 468.23 | 5.9 | 2 | 细胞外 Outside | E | GH19 |
| StuChi16 | PGSC0003DMG400016302 | chr06: 39116330-39117653 | 136 | 15 182.02 | 5.4 | 1 | 细胞质 Cytoplasm | E | GH19 |
| StuChi17 | PGSC0003DMG400021602 | chr09: 61360398-61361921 | 323 | 35 986.30 | 7.9 | 3 | 细胞外 Outside | D | GH19 |
| StuChi18 | PGSC0003DMG400025063 | chr04: 61242553-61244259 | 276 | 29 982.95 | 4.6 | 2 | 细胞外 Outside | E | GH19 |
| StuChi19 | PGSC0003DMG400025063 | chr04: 61242553-61244259 | 136 | 14 988.53 | 4.3 | 1 | 细胞质 Cytoplasm | E | GH19 |
| StuChi20 | PGSC0003DMG400026853 | chr00: 21407639-21408792 | 316 | 34 091.35 | 5.2 | 3 | 质膜Plasma membrane | F | GH19 |
| StuChi21 | PGSC0003DMG400026854 | chr00: 21415307-21416600 | 320 | 34 117.04 | 6.2 | 3 | 细胞外 Outside | F | GH19 |
| StuChi22 | PGSC0003DMG400026855 | chr00: 21419777-21420932 | 332 | 35 506.90 | 8.4 | 3 | 细胞外 Outside | F | GH19 |
| StuChi23 | PGSC0003DMG400033882 | chr05: 44304719-44306659 | 292 | 30 945.16 | 7.5 | 2 | 细胞外 Outside | C | GH18 |
| StuChi24 | PGSC0003DMG400037480 | chr07: 347182-349354 | 435 | 49 224.80 | 6.7 | 4 | 质膜Plasma membrane | A | GH18 |
| StuChi25 | PGSC0003DMG400040317 | chr00: 21299520-21300925 | 279 | 29 606.22 | 8.4 | 2 | 细胞外 Outside | F | GH19 |
| StuChi26 | PGSC0003DMG402001531 | chr02: 38760083-38761491 | 263 | 28 819.75 | 9.2 | 3 | 细胞外 Outside | F | GH19 |
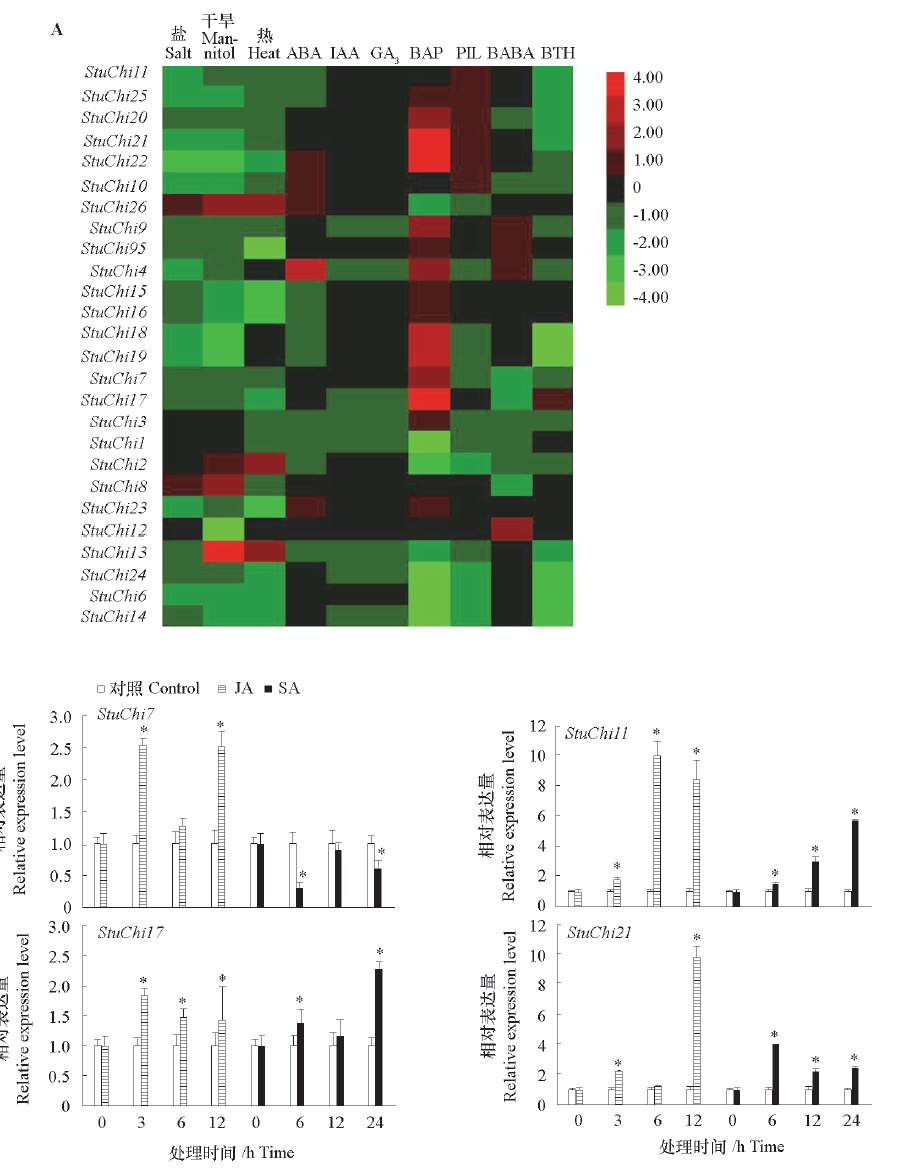
Fig. 4 Environmental stresses response analysis of StuChi genes based on RNA-Seq data(A)and exogenous hormones(B) * notes significant differences between control and treatment at 0.05 level.
| [1] |
Backiyarani S, Uma S, Nithya S, Chandrasekar A, Saraswathi M S, Thangavelu R, Mayilvaganan M, Sundararaju P, Singh N K. 2015. Genome-wide analysis and differential expression of chitinases in banana against root lesion nematode(Pratylenchus coffeae)and eumusa leaf spot (Mycosphaerella eumusae) pathogens. Applied Biochemistry and Biotechnology, 175(8):3585-3598.
doi: 10.1007/s12010-015-1528-z pmid: 25820355 |
| [2] | Bishop J G, Dean A M, Mitchell-Olds T. 2000. Rapid evolution in plant chitinases:molecular targets of selection in plant-pathogen coevolution. Proceedings of the National Academy of Sciences of the United States of America, 97(10):5322-5327. |
| [3] |
Cao J, Li X, Lv Y. 2017. Dynein light chain family genes in 15 plant species:identification,evolution and expression profiles. Plant Science, 254:70-81.
doi: 10.1016/j.plantsci.2016.10.011 URL |
| [4] | Cao J, Tan X N. 2019. Comprehensive analysis of the chitinase family genes in tomato(Solanum lycopersicum). Plants-Basel, 8(3):52. |
| [5] |
Cao R, Liu X, Gao K, Mendgen K, Kang Z, Gao J, Dai Y, Wang X. 2009. Mycoparasitism of endophytic fungi isolated from reed on soilborne phytopathogenic fungi and production of cell wall-degrading enzymes in vitro. Current Microbiology, 59(6):584-592.
doi: 10.1007/s00284-009-9477-9 URL |
| [6] |
Chen J, Piao Y, Liu Y, Li X, Piao Z. 2018. Genome-wide identification and expression analysis of chitinase gene family in Brassica rapa reveals its role in clubroot resistance. Plant Science, 270:257-267.
doi: 10.1016/j.plantsci.2018.02.017 URL |
| [7] |
Chen Q H G, Bleecker A B. 1995. Analysis of Ethylene signal-transduction kinetics associated with seedling-growth response and chitinase induction in wild-type and mutant Arabidopsis. Plant Physiology, 108(2):597-607.
doi: 10.1104/pp.108.2.597 URL |
| [8] |
Chye M L, Zhao K J, He Z M, Ramalingam S, Fung K L. 2005. An agglutinating chitinase with two chitin-binding domains confers fungal protection in transgenic potato. Planta, 220(5):717-730.
doi: 10.1007/s00425-004-1391-6 URL |
| [9] |
Cletus J, Balasubramanian V, Vashisht D, Sakthivel N. 2013. Transgenic expression of plant chitinases to enhance disease resistance. Biotechnology Letters, 35(11):1719-1732.
doi: 10.1007/s10529-013-1269-4 pmid: 23794096 |
| [10] |
Davis J M, Wu H, Cooke J E, Reed J M, Luce K S, Michler C H. 2002. Pathogen challenge,salicylic acid,and jasmonic acid regulate expression of chitinase gene homologs in pine. Molecular Plant-Microbe Interactions, 15(4):380-387.
doi: 10.1094/MPMI.2002.15.4.380 URL |
| [11] | Gregorova Z, Kovacik J, Klejdus B, Maglovski M, Kuna R, Hauptvogel P, Matusikova I. 2015. Drought-induced responses of physiology,metabolites,and PR proteins in Triticum aestivum. Journal of Agricultural & Food Chemistry, 63(37):8125-8133. |
| [12] |
Grover A. 2012. Plant chitinases:genetic diversity and physiological roles. Critical Reviews in Plant Sciences, 31:57-73.
doi: 10.1080/07352689.2011.616043 URL |
| [13] | Guo X L, Bai L R, Su C Q, Shi L R, Wang D W. 2013. Molecular cloning and expression of drought-induced protein 3(DIP3)encoding a class III chitinase in upland rice. Genetics & Molecular Research, 12(4):6860-6870. |
| [14] |
Hamel F, Bellemare G. 1995. Characterization of a class I chitinase gene and of wound-inducible,root and flower-specific chitinase expression in Brassica napus. Biochim Biophys Acta, 1263(3):212-220.
pmid: 7548207 |
| [15] |
Hou Z, Cao J. 2016. Comparative study of the P2X gene family in animals and plants. Purinergic Signal, 12(2):269-281.
doi: 10.1007/s11302-016-9501-z URL |
| [16] |
Jekel P A, Hartmann J B H, Beintema J J. 1991. The primary structure of hevamine,an enzyme with lysozyme chitinase activity from hevea-brasiliensis latex. European Journal of Biochemistry, 200(1):123-130.
pmid: 1879417 |
| [17] | Jiang C, Huang R F, Song J L, Huang M R, Xu L A. 2013. Genomewide analysis of the chitinase gene family in Populus trichocarpa. Journal of Northwest A & F University, 92(1):121-125. |
| [18] | Jiao Long, Bian Lei, Luo Zongxiu, Li Zhaoqun, Xin Zhaojun, Xiu Chunli, Cai Xiaoming, Chen Zongmao. 2020. Comparison of tea plant volatiles exogenously induced by jasmonates or salicylates elicitors. Acta Horticulturae Sinica, 47(5):927-938. (in Chinese) |
| 焦龙, 边磊, 罗宗秀, 李兆群, 辛肇军, 修春丽, 蔡晓明, 陈宗懋. 2020. 茉莉酸、水杨酸类激发子外源诱导的茶树挥发物比较. 园艺学报, 47(5):927-938. | |
| [19] |
Kashyap P, Deswal R. 2017. A novel class I Chitinase from Hippophae rhamnoides:indications for participating in ICE-CBF cold stress signaling pathway. Plant Science, 259:62-70.
doi: 10.1016/j.plantsci.2017.03.004 URL |
| [20] | Kasprzewska A. 2003. Plant chitinases-regulation and function. Cellular & Molecular Biology Letters, 8(3):809-824. |
| [21] |
Kesari P, Patil D N, Kumar P, Tomar S, Sharma A K, Kumar P. 2015. Structural and functional evolution of chitinase-like proteins from plants. Proteomics, 15(10):1693-1705.
doi: 10.1002/pmic.201400421 pmid: 25728311 |
| [22] |
Kovacs G, Sagi L, Jacon G, Arinaitwe G, Busogoro J P, Thiry E, Strosse H, Swennen R, Remy S. 2013. Expression of a rice chitinase gene in transgenic banana(‘Gros Michel’,AAA genome group)confers resistance to black leaf streak disease. Transgenic Research, 22(1):117-130.
doi: 10.1007/s11248-012-9631-1 URL |
| [23] |
Lacombe-Harvey M E, Brzezinski R, Beaulieu C. 2018. Chitinolytic functions in actinobacteria:ecology,enzymes,and evolution. Applied Microbiology and Biotechnology, 102(17):7219-7230.
doi: 10.1007/s00253-018-9149-4 pmid: 29931600 |
| [24] |
Lazar A, Coll A, Dobnik D, Baebler S, Bedina-Zavec A, Zel J, Gruden K. 2014. Involvement of potato(Solanum tuberosum L.)MKK6 in response to Potato virus Y. PLoS ONE, 9(8):e104553.
doi: 10.1371/journal.pone.0104553 URL |
| [25] |
Lee C G, Da Silva C A, Dela Cruz C S, Ahangari F, Ma B, Kang M J, He C H, Takyar S, Elias J A. 2011. Role of chitin and chitinase/chitinase-like proteins in inflammation,tissue remodeling,and injury. Annual Review of Physiology, 73:479-501.
doi: 10.1146/annurev-physiol-012110-142250 URL |
| [26] |
Lee J H, Takei K, Sakakibara H, Cho H S, Kim D M, Kim Y S, Min S R, Kim W T, Sohn D Y, Lim Y P, Pai H S. 2003. CHRK1,a chitinase-related receptor-like kinase,plays a role in plant development and cytokinin homeostasis in tobacco. Plant Molecular Biology, 53(6):877-890.
doi: 10.1023/B:PLAN.0000023668.34205.a8 URL |
| [27] |
Li H, Greene L H. 2010. Sequence and structural analysis of the chitinase insertion domain reveals two conserved motifs involved in chitin-binding. PLoS ONE, 5(1):e8654.
doi: 10.1371/journal.pone.0008654 URL |
| [28] | Li Qiang, Qi Jingjing, Dou Wanfu, Qin Xiujuan, He Yongrui, Chen Shanchun. 2020. Overexpression of CsNBS-LRR in citrus confers bacterial canker resistance by regulating SA signaling pathway. Acta Horticulturae Sinica, 47(5):817-826. (in Chinese) |
| 李强, 祁静静, 窦万福, 秦秀娟, 何永睿, 陈善春. 2020. 柑橘超量表达CsNBS-LRR通过SA信号途径增强对溃疡病抗性. 园艺学报, 47(5):817-826. | |
| [29] | Lin W, Anuratha C S, Datta K, Potrykus I, Muthukrishnan S, Datta S K. 1995. Genetic-engineering of rice for resistance to sheath blight. Bio-Technology, 13(7):686-691. |
| [30] |
Liu Y, Wang L, Xing X, Sun L, Pan J, Kong X, Zhang M, Li D. 2013. ZmLEA3,a multifunctional group 3 LEA protein from maize(Zea mays L.),is involved in biotic and abiotic stresses. Plant and Cell Physiology, 54(6):944-959.
doi: 10.1093/pcp/pct047 URL |
| [31] | Liu Xia, Huang Xxun, Du Xia, Zhao Bin, Yang Yanli. 2019. The effect of salicylic acid on late blight resistance of different potato varieties. Enshi:China Potato Conference. (in Chinese) |
| 刘霞, 黄勋, 杜霞, 赵彬, 杨艳丽. 2019. 水杨酸对不同马铃薯品种晚疫病抗性的影响. 恩施:中国马铃薯大会. | |
| [32] |
Lopez R C, Gomez-Gomez L. 2009. Isolation of a new fungi and wound-induced chitinase class in corms of Crocus sativus. Plant Physiology and Biochemistry, 47(5):426-434.
doi: 10.1016/j.plaphy.2009.01.007 URL |
| [33] |
Ma H R, Wang F, Wang W J, Yin G Y, Zhang D Y, Ding Y Q, Timko M P, Zhang H B. 2016. Alternative splicing of basic chitinase gene PR3b in the low-nicotine mutants of Nicotiana tabacum L. cv. Burley 21. Journal of Experimental Botany, 67(19):5799-5809.
doi: 10.1093/jxb/erw345 URL |
| [34] |
Malolepszy A, Kelly S, Sorensen K K, James E K, Kalisch C, Bozsoki Z, Panting M, Andersen S U, Sato S, Tao K, Jesen D B, Vinter M, de Jong N, Madsen L H, Vmehara Y, Gysel K, Berentsen M V, Blaise M, Jensen K J, Thygesen M B, Sandal N, Andersen K R, Radutoiu S. 2018. A plant chitinase controls cortical infection thread progression and nitrogen-fixing symbiosis. Elife, 7:e38874.
doi: 10.7554/eLife.38874 URL |
| [35] |
Maksimov I V, Abizgildina P P, Sorokan’A V, Burkhanova G F. 2014. Regulation of peroxidase activity under the influence of signaling molecules and Bacillus subtilis 26D in potato plants infected with Phytophthora infestans. Applied Biochemistry and Microbiology, 50(2):173-8
doi: 10.1134/S0003683814020136 URL |
| [36] |
Margispinheiro M, Metzboutigue M H, Awade A, Detapia M, Leret M, Burkard G. 1991. Isolation of a complementary-DNA encoding the bean Pr4 chitinase-an acidic enzyme with an amino-terminus cysteine-rich domain. Plant Molecular Biology, 17(2):243-253.
doi: 10.1007/BF00039499 URL |
| [37] |
Neuhaus J M, Fritig B, Linthorst H J M, Meins F, Mikkelsen J D, Ryals J. 1996. A revised nomenclature for chitinase genes. Plant Molecular Biology Reporter, 14(2):102-104.
doi: 10.1007/BF02684897 URL |
| [38] | Nishizawa Y, Nishio Z, Nakazono K, Soma M, Nakajima E, Ugaki M, Hibi T. 1999. Enhanced resistance to blast(Magnaporthe grisea)in transgenic Japonica rice by constitutive expression of rice chitinase. Theoretical & Applied Genetics, 99(3-4):383-390. |
| [39] |
Ohnuma T, Sorlie M, Fukuda T, Kawamoto N, Taira T, Fukamizo T. 2011. Chitin oligosaccharide binding to a family GH19 chitinase from the moss Bryum coronatum. The Febs Journal, 278(21):3991-4001.
doi: 10.1111/j.1742-4658.2011.08301.x URL |
| [40] |
Passarinho P A, Van Hengel A J, Fransz P F, de Vries S C. 2001. Expression pattern of the Arabidopsis thaliana AtEP3/Atchit IV endochitinase gene. Planta, 212(4):556-567.
pmid: 11525512 |
| [41] |
Petit A N, Baillieul F, Vaillant-Gaveau N, Jacquens L, Conreux A, Jeandet P, Clement C, Fontaine F. 2009. Low responsiveness of grapevine flowers and berries at fruit set to UV-C irradiation. Journal of Experimental Botany, 60(4):1155-1162.
doi: 10.1093/jxb/ern361 URL |
| [42] | Punja Z K, Zhang Y Y. 1993. Plant chitinases and their roles in resistance to fungal diseases. Journal of Nematology, 25(4):526-540. |
| [43] | Ren Yajuan, Wang Haixia, Li Yajun, Chen Yanlin, Wang Jing, Tian Zhendong. 2015. An early β-a minobutyric acid responsive gene StWRKY5 confers resistance to late blight in potato. Molecular Plant Breeding,(6):1207-1213. (in Chinese) |
| 任亚娟, 王海霞, 李亚军, 陈艳林, 王静, 田振东. 2015. β-氨基丁酸早期诱导表达基因StWRKY5参与马铃薯晚疫病抗性. 分子植物育种,(6):1207-1213. | |
| [44] |
Renner T, Specht C D. 2012. Molecular and functional evolution of class I chitinases for plant carnivory in the caryophyllales. Molecular Biology and Evolution, 29(10):2971-2985.
pmid: 22490823 |
| [45] |
Shinya T, Hanai K, Galis I, Suzuki K, Matsuoka K, Matsuoka H, Saito M. 2007. Characterization of NtChitIV,a class IV chitinase induced by beta-1,3-,1,6-glucan elicitor from Alternaria alternata 102:antagonistic effect of salicylic acid and methyl jasmonate on the induction of NtChitIV. Biochemical and Biophysical Research Communications, 353(2):311-317.
doi: 10.1016/j.bbrc.2006.12.009 URL |
| [46] |
Shoresh M, Harman G E. 2008. Genome-wide identification,expression and chromosomal location of the genes encoding chitinolytic enzymes in Zea mays. Molecular Genetics and Genomics, 280(2):173-185.
doi: 10.1007/s00438-008-0354-1 URL |
| [47] |
Su Y, Xu L, Fu Z, Yang Y, Guo J, Wang S, Que Y. 2014. ScChi,encoding an acidic class III chitinase of sugarcane,confers positive responses to biotic and abiotic stresses in sugarcane. International Journal of Molecular Sciences, 15(2):2738-2760.
doi: 10.3390/ijms15022738 URL |
| [48] |
Su Y, Xu L, Wang S, Wang Z, Yang Y, Chen Y, Que Y. 2015. Identification,phylogeny,and transcript of chitinase family genes in sugarcane. Scientific Reports, 5:10708.
doi: 10.1038/srep10708 URL |
| [49] |
Suginta W, Sirimontree P, Sritho N, Ohnuma T, Fukamizo T. 2016. The chitin-binding domain of a GH-18 chitinase from Vibrio harveyi is crucial for chitin-chitinase interactions. International Journal of Biological Macromolecules, 93:1111-1117.
doi: S0141-8130(16)31678-6 pmid: 27667544 |
| [50] | Takashima T, Numata T, Taira T, Fukamizo T, Ohnuma T. 2018. Structure and enzymatic properties of a two-domain family GH19 chitinase from Japanese cedar(Cryptomeria japonica) pollen. Journal of Agricultural & Food Chemistry, 66(22):5699-5706. |
| [51] |
Takenaka Y, Nakano S, Tamoi M, Sakuda S, Fukamizo T. 2009. Chitinase gene expression in response to environmental stresses in Arabidopsis thaliana:chitinase inhibitor allosamidin enhances stress tolerance. Biosci Biotechnol Biochem, 73(5):1066-1071.
doi: 10.1271/bbb.80837 URL |
| [52] |
van der Holst P P, Schlaman H R, Spaink H P. 2001. Proteins involved in the production and perception of oligosaccharides in relation to plant and animal development. Current Opinion in Structural Biology, 11(5):608-616.
doi: 10.1016/S0959-440X(00)00255-4 URL |
| [53] |
Wang L Y, Wang Y S, Cheng H, Zhang J P, Yeok F S. 2015. Cloning of the Aegiceras corniculatum class I chitinase gene(AcCHII)and the response of AcCHII mRNA expression to cadmium stress. Ecotoxicology, 24(7-8):1705-1713.
doi: 10.1007/s10646-015-1502-0 URL |
| [54] |
White R F, Rybicki E P, Vonwvhmar M B, Dekker J L, Antoniw J F. 1987. Detection of PR-1 type proteins in Amaranthaceae,Cheuopodiaceae,Graminae and Solanaceae by immunoelectroblotting. Journal of General Virology, 68:2043-2048.
doi: 10.1099/0022-1317-68-7-2043 URL |
| [55] |
Wu J, Wang Y, Kim S T, Kim S G, Kang K Y. 2013. Characterization of a newly identified rice chitinase-like protein(OsCLP)homologous to xylanase inhibitor. BMC Biotechnology, 13:4.
doi: 10.1186/1472-6750-13-4 URL |
| [56] |
Xu F, Fan C, He Y. 2007. Chitinases in Oryza sativa ssp. japonica and Arabidopsis thaliana. Journal of Genetics and Genomics, 34(2):138-150.
doi: 10.1016/S1673-8527(07)60015-0 URL |
| [57] | Xu J, Xu X, Tian L, Wang G, Zhang X, Wang X, Guo W. 2016. Discovery and identification of candidate genes from the chitinase gene family for Verticillium dahliae resistance in cotton. Science Reports, 6:29022. |
| [58] | Yang J, Zhang K Q. 2019. Chitin synthesis and degradation in fungi biology and enzymes. Advances in Experimental Medicine and Biology, 1142:153-167. |
| [59] | Yang Yanli. 2010. Preliminary study of exogenous jasmonic acid effects on potato resistance to late blight[Ph. D. Dissertation]. Changsha:Hunan Agricultural University. (in Chinese) |
| 杨艳丽. 2010. 外源茉莉酸影响马铃薯对晚疫病抗性的初步研究[博士论文]. 长沙:湖南农业大学. | |
| [60] |
Zhang C, Zhang L, Wang D, Ma H, Liu B, Shi Z, Ma X H, Chen Y, Chen Q. 2018. Evolutionary history of the glycoside hydrolase 3(GH3)family based on the sequenced genomes of 48 plants and identification of jasmonic acid-related GH3 proteins in Solanum tuberosum. International Journal of Molecular Sciences, 19(7):1850.
doi: 10.3390/ijms19071850 URL |
| [1] | WANG Xiaochen, NIE Ziye, LIU Xianju, DUAN Wei, FAN Peige, and LIANG Zhenchang, . Effects of Abscisic Acid on Monoterpene Synthesis in‘Jingxiangyu’Grape Berries [J]. Acta Horticulturae Sinica, 2023, 50(2): 237-249. |
| [2] | WANG Rui, HONG Wenjuan, LUO Hua, ZHAO Lina, CHEN Ying, and WANG Jun, . Construction of SSR Fingerprints of Pomegranate Cultivars and Male Parent Identification of Hybrids [J]. Acta Horticulturae Sinica, 2023, 50(2): 265-278. |
| [3] | ZHANG Xin, QI Yanxiang, ZENG Fanyun, WANG Yanwei, XIE Peilan, XIE Yixian, and PENG Jun. Functional Analysis of Dicer-like Genes in Fusarium oxysporum f. sp. cubense Race 4 [J]. Acta Horticulturae Sinica, 2023, 50(2): 279-294. |
| [4] | YU Tingting, LI Huan, NING Yuansheng, SONG Jianfei, PENG Lulin, JIA Junqi, ZHANG Weiwei, and YANG Hongqiang. Genome-wide Identification of GRAS Gene Family in Apple and Expression Analysis of Its Response to Auxin [J]. Acta Horticulturae Sinica, 2023, 50(2): 397-409. |
| [5] | LIANG Jiali, WU Qisong, CHEN Guangquan, ZHANG Rong, XU Chunxiang, and FENG Shujie, . Identification of the Neopestalotiopsis musae Pathogen of Banana Leaf Spot Disease [J]. Acta Horticulturae Sinica, 2023, 50(2): 410-420. |
| [6] | WANG Mengmeng, SUN Deling, CHEN Rui, YANG Yingxia, ZHANG Guan, LÜ Mingjie, WANG Qian, XIE Tianyu, NIU Guobao, SHAN Xiaozheng, TAN Jin, and YAO Xingwei, . Construction and Evaluation of Cauliflower Core Collection [J]. Acta Horticulturae Sinica, 2023, 50(2): 421-431. |
| [7] | WANG Xiqing, JIA Yunhe, YAN Wen, FU Yongkai, YOU Haibo, LI Dongyan, and ZHAO Jingchao. A New Watermelon Cultivar‘Longsheng Jiali’with High Resistance to Fusarium Wilt [J]. Acta Horticulturae Sinica, 2023, 50(2): 455-456. |
| [8] | ZHAI Hanhan, ZHAI Yujie, TIAN Yi, ZHANG Ye, YANG Li, WEN Zhiliang, CHEN Haijiang. Genome-wide Identification of Peach SAUR Gene Family and Characterization of PpSAUR5 Gene [J]. Acta Horticulturae Sinica, 2023, 50(1): 1-14. |
| [9] | YANG Zhi, ZHANG Chuanjiang, YANG Xinfang, DONG Mengyi, WANG Zhenlei, YAN Fenfen, WU Cuiyun, WANG Jiurui, LIU Mengjun, LIN Minjuan. Analysis of Fruit Genetic Tendency and Mixed Inheritance in Hybrid Progeny of Jujube and Wild Jujube [J]. Acta Horticulturae Sinica, 2023, 50(1): 36-52. |
| [10] | HU Jingyu, QUE Kaijuan, MIAO Tianli, WU Shaozheng, WANG Tiantian, ZHANG Lei, DONG Xian, JI Pengzhang, DONG Jiahong. Identification of Tomato Spotted Wilt Orthotospovirus Infecting Iris tectorum [J]. Acta Horticulturae Sinica, 2023, 50(1): 170-176. |
| [11] | ZHAO Xueyan, WANG Qi, WANG Li, WANG Fangyuan, WANG Qing, LI Yan. Comparative Transcriptome Analysis of Differential Expression in Different Tissues of Corydalis yanhusuo [J]. Acta Horticulturae Sinica, 2023, 50(1): 177-187. |
| [12] | SHAO Fengqing, LUO Xiurong, WANG Qi, ZHANG Xianzhi, WANG Wencai. Advances in Research of DNA Methylation Regulation During Fruit Ripening [J]. Acta Horticulturae Sinica, 2023, 50(1): 197-208. |
| [13] | YU Yangjun, SU Tongbing, ZHANG Fenglan, ZHANG Deshuang, ZHAO Xiuyun, YU Shuancang, WANG Weihong, LI Peirong, XIN Xiaoyun, WANG Jiao, and WU Changjian. A New Purple Seedling-Edible Chinese Cabbage F1 Hybrid‘Jingyan Zikuaicai’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 91-92. |
| [14] | LI Zhengli, ZHANG Li, and MA Licang. A New Capsicum frutescens Cultivar‘Huangla Chaotian’with Yellow Fruit [J]. Acta Horticulturae Sinica, 2022, 49(S2): 123-124. |
| [15] | WANG Kuan, QI Lipan, WU Guili, FENG Yan, WANG Lei, YIN Jiang, LUO Yating, WANG Yan, LIU Chang, GONG Xuechen, and WANG Haijun. A New Potato Cultivar‘Beifang 002’of Early Maturity and Good Yield [J]. Acta Horticulturae Sinica, 2022, 49(S2): 141-142. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd