
园艺学报 ›› 2023, Vol. 50 ›› Issue (6): 1230-1242.doi: 10.16420/j.issn.0513-353x.2022-0509
黄治皓1, 刘婷婷1, 董旭杰1, 严明理2, 刘志祥1, 曾超珍1,*( )
)
收稿日期:2022-10-21
修回日期:2023-03-14
出版日期:2023-06-25
发布日期:2023-06-27
通讯作者:
* (E-mail:chaozhenzeng@163.com)基金资助:
HUANG Zhihao1, LIU Tingting1, DONG Xujie1, YAN Mingli2, LIUZhixiang 1, ZENG Chaozhen1,*( )
)
Received:2022-10-21
Revised:2023-03-14
Published:2023-06-25
Online:2023-06-27
摘要:
芥菜(Brassica juncea)对多种重金属具有富集能力。重金属ATP酶(heavy metal transporting ATPase,HMA)在植物转运重金属过程中具有重要的作用。基于基因组和转录组数据,对芥菜HMA家族基因成员进行了全基因组鉴定。芥菜基因组中包含27个HMA基因,其编码的蛋白质中有6个不稳定指数大于40,有22个为疏水性蛋白;这些基因聚类为P-1B-1、P-1B-2和P-1B-4等3个亚家族;27个HMA蛋白均为膜蛋白,都具有E1-E2 ATPase和hydrolase结构域;芥菜HMA蛋白都含有8 ~ 15个保守基序,不同亚族因具有独特的保守基序结构从而转运重金属的种类不同;在芥菜HMA基因启动子区域中与生长发育有关的顺式作用元件TGA和与逆境响应相关的顺式作用元件ABRE、MBS、LTR、TC广泛分布;在镉胁迫下,BjuA033764、BjuB019118、BjuB035256、BjuB045293等基因在叶片中表达量明显上调,BjuA003596、BjuB040613、BjuA025022等基因在根系中表达量明显上调,说明其参与了芥菜对镉胁迫的响应。
中图分类号:
黄治皓, 刘婷婷, 董旭杰, 严明理, 刘志祥, 曾超珍. 芥菜HMA家族基因鉴定及其在镉胁迫下的表达分析[J]. 园艺学报, 2023, 50(6): 1230-1242.
HUANG Zhihao, LIU Tingting, DONG Xujie, YAN Mingli, LIUZhixiang , ZENG Chaozhen. Identification and Expression Analysis of HMA Gene Family in Brassica juncea Under Cadmium Stress[J]. Acta Horticulturae Sinica, 2023, 50(6): 1230-1242.
| 基因ID Gene ID | 正向引物(5′-3′) Forward primer | 反向引物(5′-3′) Reverse primer |
|---|---|---|
| BjuB002000 | ATCGTTGGCTTCCCTCTCGTT | ACATCCACCATTGACCGGCTTG |
| BjuA003596 | AAGAACCGTCATCGTCGTCCA | AGAAGTACGCCGGAAACCAC |
| BjuB040613 | TCATCGTCGTCCATGATAGCC | AATTAGTTTCTCCGGTTACCCTC |
| BjuO003139 | CGATACTTGCCAAAGCCGTTG | TGCATAACCGCATTCGCCTT |
| BjuA025267 | GCCTCCTCATCTCTCCGTCCC | TTTGCCGTCCACTTTCACGTT |
| BjuA042408 | CAAGAACCGTCATCGTTGTCC | GCAACAAGAGCGAACCACT |
| Actin | GAATCCACGAGACGACTTACAAC | CGATCCAGACACTGTACTTCCTC |
表1 qRT-PCR引物序列
Table 1 The sequences of qRT-PCR primers
| 基因ID Gene ID | 正向引物(5′-3′) Forward primer | 反向引物(5′-3′) Reverse primer |
|---|---|---|
| BjuB002000 | ATCGTTGGCTTCCCTCTCGTT | ACATCCACCATTGACCGGCTTG |
| BjuA003596 | AAGAACCGTCATCGTCGTCCA | AGAAGTACGCCGGAAACCAC |
| BjuB040613 | TCATCGTCGTCCATGATAGCC | AATTAGTTTCTCCGGTTACCCTC |
| BjuO003139 | CGATACTTGCCAAAGCCGTTG | TGCATAACCGCATTCGCCTT |
| BjuA025267 | GCCTCCTCATCTCTCCGTCCC | TTTGCCGTCCACTTTCACGTT |
| BjuA042408 | CAAGAACCGTCATCGTTGTCC | GCAACAAGAGCGAACCACT |
| Actin | GAATCCACGAGACGACTTACAAC | CGATCCAGACACTGTACTTCCTC |
| 基因Gene | 结构域Domain | 基因Gene | 结构域Domain | |||||
|---|---|---|---|---|---|---|---|---|
| HMA | E1-E2 ATPase | Hydrolase | HMA | E1-E2 ATPase | Hydrolase | |||
| AtHMA1 | - | E1-E2 ATPase | Hydrolase | BjuO003139 | - | E1-E2 ATPase | Hydrolase | |
| AtHMA2 | - | E1-E2 ATPase | Hydrolase | BjuB035256 | - | E1-E2 ATPase | Hydrolase | |
| AtHMA3 | - | E1-E2 ATPase | Hydrolase | BjuA023298 | - | E1-E2 ATPase | Hydrolase | |
| AtHMA4 | - | E1-E2 ATPase | Hydrolase | BjuA025267 | - | E1-E2 ATPase | Hydrolase | |
| AtHMA5 | [HMA]3 | E1-E2 ATPase | Hydrolase | BjuA042408 | HMA | E1-E2 ATPase | Hydrolase | |
| AtHMA6 | HMA | E1-E2 ATPase | Hydrolase | BjuB028207 | [HMA]2 | E1-E2 ATPase | Hydrolase | |
| AtHMA7 | [HMA]2 | E1-E2 ATPase | Hydrolase | BjuA046861 | [HMA]2 | E1-E2 ATPase | Hydrolase | |
| AtHMA8 | HMA | E1-E2 ATPase | Hydrolase | BjuB035323 | [HMA]2 | E1-E2 ATPase | Hydrolase | |
| BjuB002000 | - | E1-E2 ATPase | Hydrolase | BjuA038121 | [HMA]2 | E1-E2 ATPase | Hydrolase | |
| BjuB034828 | - | E1-E2 ATPase | Hydrolase | BjuB019118 | HMA | E1-E2 ATPase | Hydrolase | |
| BjuA036645 | - | E1-E2 ATPase | Hydrolase | BjuB027969 | [HMA]2 | E1-E2 ATPase | Hydrolase | |
| BjuA002855 | - | E1-E2 ATPase | Hydrolase | BjuA025022 | [HMA]2 | E1-E2 ATPase | Hydrolase | |
| BjuO011640 | HMA | E1-E2 ATPase | Hydrolase | BjuB036631 | [HMA]2 | E1-E2 ATPase | Hydrolase | |
| BjuA003595 | - | E1-E2 ATPase | Hydrolase | BjuA033764 | [HMA]2 | E1-E2 ATPase | Hydrolase | |
| BjuB040614 | - | E1-E2 ATPase | Hydrolase | BjuA003262 | HMA | E1-E2 ATPase | Hydrolase | |
| BjuA003596 | HMA | E1-E2 ATPase | Hydrolase | BjuA038847 | HMA | E1-E2 ATPase | Hydrolase | |
| BjuB040613 | HMA | E1-E2 ATPase | Hydrolase | BjuB045293 | HMA | E1-E2 ATPase | Hydrolase | |
| BjuA042404 | [HMA]3 | E1-E2 ATPase | Hydrolase | |||||
表2 HMA家族基因的结构域分布
Table 2 Distribution of domains in HMA gene family
| 基因Gene | 结构域Domain | 基因Gene | 结构域Domain | |||||
|---|---|---|---|---|---|---|---|---|
| HMA | E1-E2 ATPase | Hydrolase | HMA | E1-E2 ATPase | Hydrolase | |||
| AtHMA1 | - | E1-E2 ATPase | Hydrolase | BjuO003139 | - | E1-E2 ATPase | Hydrolase | |
| AtHMA2 | - | E1-E2 ATPase | Hydrolase | BjuB035256 | - | E1-E2 ATPase | Hydrolase | |
| AtHMA3 | - | E1-E2 ATPase | Hydrolase | BjuA023298 | - | E1-E2 ATPase | Hydrolase | |
| AtHMA4 | - | E1-E2 ATPase | Hydrolase | BjuA025267 | - | E1-E2 ATPase | Hydrolase | |
| AtHMA5 | [HMA]3 | E1-E2 ATPase | Hydrolase | BjuA042408 | HMA | E1-E2 ATPase | Hydrolase | |
| AtHMA6 | HMA | E1-E2 ATPase | Hydrolase | BjuB028207 | [HMA]2 | E1-E2 ATPase | Hydrolase | |
| AtHMA7 | [HMA]2 | E1-E2 ATPase | Hydrolase | BjuA046861 | [HMA]2 | E1-E2 ATPase | Hydrolase | |
| AtHMA8 | HMA | E1-E2 ATPase | Hydrolase | BjuB035323 | [HMA]2 | E1-E2 ATPase | Hydrolase | |
| BjuB002000 | - | E1-E2 ATPase | Hydrolase | BjuA038121 | [HMA]2 | E1-E2 ATPase | Hydrolase | |
| BjuB034828 | - | E1-E2 ATPase | Hydrolase | BjuB019118 | HMA | E1-E2 ATPase | Hydrolase | |
| BjuA036645 | - | E1-E2 ATPase | Hydrolase | BjuB027969 | [HMA]2 | E1-E2 ATPase | Hydrolase | |
| BjuA002855 | - | E1-E2 ATPase | Hydrolase | BjuA025022 | [HMA]2 | E1-E2 ATPase | Hydrolase | |
| BjuO011640 | HMA | E1-E2 ATPase | Hydrolase | BjuB036631 | [HMA]2 | E1-E2 ATPase | Hydrolase | |
| BjuA003595 | - | E1-E2 ATPase | Hydrolase | BjuA033764 | [HMA]2 | E1-E2 ATPase | Hydrolase | |
| BjuB040614 | - | E1-E2 ATPase | Hydrolase | BjuA003262 | HMA | E1-E2 ATPase | Hydrolase | |
| BjuA003596 | HMA | E1-E2 ATPase | Hydrolase | BjuA038847 | HMA | E1-E2 ATPase | Hydrolase | |
| BjuB040613 | HMA | E1-E2 ATPase | Hydrolase | BjuB045293 | HMA | E1-E2 ATPase | Hydrolase | |
| BjuA042404 | [HMA]3 | E1-E2 ATPase | Hydrolase | |||||
| 亚族 Subgroup | 基因 Gene | 长度/aa Length | 分子量/kD MW | 等电点 pI | 不稳定指数Instability index | 疏水性指数 GRAVY | 亚细胞定位 Subcellular location | |
|---|---|---|---|---|---|---|---|---|
| WoLF PSORT | BUSCA | |||||||
| P-1B-1 | BjuB028207 | 998 | 108.48 | 5.72 | 35.69 | 0.16 | PM | PM |
| BjuA046861 | 1 184 | 129.44 | 5.62 | 35.11 | 0.08 | PM | PM | |
| BjuB019118 | 839 | 90.81 | 5.55 | 27.34 | 0.28 | PM | PM | |
| BjuB035323 | 978 | 106.55 | 5.94 | 28.84 | 0.17 | PM | PM | |
| BjuA038121 | 973 | 105.65 | 6.13 | 26.97 | 0.19 | PM | PM | |
| BjuB027969 | 1 006 | 107.54 | 5.02 | 32.02 | 0.22 | PM | PM | |
| BjuA033764 | 980 | 104.40 | 4.93 | 31.54 | 0.23 | PM | PM | |
| BjuA025022 | 999 | 106.84 | 5.00 | 33.47 | 0.25 | PM | PM | |
| BjuB036631 | 1 000 | 106.96 | 5.01 | 33.48 | 0.25 | PM | PM | |
| BjuA003262 | 938 | 98.60 | 8.76 | 41.58 | 0.11 | PM | MM | |
| BjuA038847 | 885 | 94.22 | 5.68 | 37.54 | 0.16 | PM | MM | |
| P-1B-2 | BjuA002855 | 683 | 73.81 | 7.23 | 33.38 | 0.06 | PM | MM |
| BjuO011640 | 801 | 86.29 | 7.10 | 36.86 | 0.22 | PM | PM | |
| BjuA003595 | 758 | 81.39 | 5.98 | 33.75 | 0.26 | PM | PM | |
| BjuB040614 | 760 | 81.84 | 5.88 | 33.83 | 0.25 | PM | PM | |
| BjuA003596 | 872 | 94.33 | 6.93 | 35.83 | 0.04 | PM | PM | |
| BjuB040613 | 928 | 99.86 | 6.78 | 35.05 | 0.04 | PM | PM | |
| BjuA042404 | 903 | 97.14 | 7.97 | 32.33 | 0 | PM | PM | |
| BjuO003139 | 1 271 | 137.78 | 7.79 | 84.05 | −0.24 | PM | PM | |
| BjuB035256 | 1 220 | 132.84 | 6.23 | 43.35 | −0.19 | PM | PM | |
| BjuA023298 | 1 193 | 129.78 | 6.82 | 41.95 | −0.23 | PM | PM | |
| BjuA025267 | 1 162 | 125.17 | 6.31 | 91.34 | −0.08 | PM | PM | |
| BjuA042408 | 795 | 85.53 | 6.80 | 38.11 | 0.25 | PM | PM | |
| BjuB045293 | 613 | 63.83 | 9.29 | 46.64 | 0.22 | PM | MM | |
| P-1B-4 | BjuB002000 | 820 | 88.05 | 7.87 | 35.75 | 0.12 | PM | MM |
| BjuB034828 | 783 | 84.77 | 6.86 | 35.14 | 0.18 | PM | OM | |
| BjuA036645 | 775 | 84.25 | 6.74 | 34.78 | 0.13 | PM | OM | |
表3 HMA家族的蛋白质信息和亚细胞定位
Table 3 Protein information and subcellular localization of HMA family
| 亚族 Subgroup | 基因 Gene | 长度/aa Length | 分子量/kD MW | 等电点 pI | 不稳定指数Instability index | 疏水性指数 GRAVY | 亚细胞定位 Subcellular location | |
|---|---|---|---|---|---|---|---|---|
| WoLF PSORT | BUSCA | |||||||
| P-1B-1 | BjuB028207 | 998 | 108.48 | 5.72 | 35.69 | 0.16 | PM | PM |
| BjuA046861 | 1 184 | 129.44 | 5.62 | 35.11 | 0.08 | PM | PM | |
| BjuB019118 | 839 | 90.81 | 5.55 | 27.34 | 0.28 | PM | PM | |
| BjuB035323 | 978 | 106.55 | 5.94 | 28.84 | 0.17 | PM | PM | |
| BjuA038121 | 973 | 105.65 | 6.13 | 26.97 | 0.19 | PM | PM | |
| BjuB027969 | 1 006 | 107.54 | 5.02 | 32.02 | 0.22 | PM | PM | |
| BjuA033764 | 980 | 104.40 | 4.93 | 31.54 | 0.23 | PM | PM | |
| BjuA025022 | 999 | 106.84 | 5.00 | 33.47 | 0.25 | PM | PM | |
| BjuB036631 | 1 000 | 106.96 | 5.01 | 33.48 | 0.25 | PM | PM | |
| BjuA003262 | 938 | 98.60 | 8.76 | 41.58 | 0.11 | PM | MM | |
| BjuA038847 | 885 | 94.22 | 5.68 | 37.54 | 0.16 | PM | MM | |
| P-1B-2 | BjuA002855 | 683 | 73.81 | 7.23 | 33.38 | 0.06 | PM | MM |
| BjuO011640 | 801 | 86.29 | 7.10 | 36.86 | 0.22 | PM | PM | |
| BjuA003595 | 758 | 81.39 | 5.98 | 33.75 | 0.26 | PM | PM | |
| BjuB040614 | 760 | 81.84 | 5.88 | 33.83 | 0.25 | PM | PM | |
| BjuA003596 | 872 | 94.33 | 6.93 | 35.83 | 0.04 | PM | PM | |
| BjuB040613 | 928 | 99.86 | 6.78 | 35.05 | 0.04 | PM | PM | |
| BjuA042404 | 903 | 97.14 | 7.97 | 32.33 | 0 | PM | PM | |
| BjuO003139 | 1 271 | 137.78 | 7.79 | 84.05 | −0.24 | PM | PM | |
| BjuB035256 | 1 220 | 132.84 | 6.23 | 43.35 | −0.19 | PM | PM | |
| BjuA023298 | 1 193 | 129.78 | 6.82 | 41.95 | −0.23 | PM | PM | |
| BjuA025267 | 1 162 | 125.17 | 6.31 | 91.34 | −0.08 | PM | PM | |
| BjuA042408 | 795 | 85.53 | 6.80 | 38.11 | 0.25 | PM | PM | |
| BjuB045293 | 613 | 63.83 | 9.29 | 46.64 | 0.22 | PM | MM | |
| P-1B-4 | BjuB002000 | 820 | 88.05 | 7.87 | 35.75 | 0.12 | PM | MM |
| BjuB034828 | 783 | 84.77 | 6.86 | 35.14 | 0.18 | PM | OM | |
| BjuA036645 | 775 | 84.25 | 6.74 | 34.78 | 0.13 | PM | OM | |
| 亚族 Subgroup | 基因 Gene | 染色体 Chr | 顺式作用元件数量 Number of cis-acting element | |||||
|---|---|---|---|---|---|---|---|---|
| G-Box | ABRE | LTR | TC | MBS | TGA | |||
| P-1B-1 | AtHMA5 | 1 | 1 | 1 | 0 | 2 | 0 | 2 |
| AtHMA6 | 4 | 0 | 1 | 0 | 1 | 1 | 1 | |
| AtHMA7 | 5 | 1 | 1 | 0 | 3 | 1 | 1 | |
| AtHMA8 | 5 | 0 | 1 | 0 | 1 | 2 | 1 | |
| BjuB028207 | B04 | 0 | 1 | 0 | 0 | 0 | 1 | |
| BjuA046861 | A09 | 0 | 0 | 0 | 0 | 0 | 0 | |
| BjuB019118 | B08 | 0 | 0 | 0 | 0 | 1 | 0 | |
| BjuB035323 | B02 | 0 | 0 | 0 | 1 | 0 | 0 | |
| BjuA038121 | A10 | 0 | 0 | 0 | 2 | 0 | 0 | |
| BjuB027969 | B04 | 0 | 0 | 1 | 3 | 1 | 0 | |
| BjuA033764 | A09 | 0 | 0 | 1 | 3 | 1 | 0 | |
| BjuA025022 | A06 | 1 | 1 | 0 | 0 | 1 | 0 | |
| BjuB036631 | B02 | 1 | 1 | 0 | 0 | 1 | 0 | |
| BjuA003262 | A01 | 0 | 0 | 1 | 0 | 0 | 0 | |
| BjuA038847 | A10 | 7 | 7 | 2 | 0 | 1 | 0 | |
| P-1B-2 | AtHMA2 | 4 | 1 | 0 | 0 | 1 | 2 | 0 |
| AtHMA3 | 4 | 1 | 0 | 1 | 0 | 2 | 2 | |
| AtHMA4 | 2 | 0 | 1 | 1 | 1 | 0 | 3 | |
| BjuA002855 | A01 | 0 | 0 | 0 | 0 | 1 | 0 | |
| BjuO011640 | Super_scaffold_184_753382_1581011 | 1 | 1 | 0 | 0 | 1 | 2 | |
| BjuA003595 | A01 | 0 | 0 | 0 | 0 | 0 | 0 | |
| BjuB040614 | B05 | 1 | 1 | 0 | 2 | 0 | 0 | |
| BjuA003596 | A01 | 2 | 2 | 2 | 1 | 0 | 3 | |
| BjuB040613 | B05 | 2 | 2 | 0 | 0 | 0 | 0 | |
| BjuA042404 | A03 | 2 | 3 | 2 | 0 | 1 | 0 | |
| BjuO003139 | Contig2023 | 2 | 2 | 1 | 0 | 0 | 0 | |
| BjuB035256 | B02 | 3 | 1 | 3 | 1 | 0 | 1 | |
| BjuA023298 | A06 | 1 | 1 | 0 | 2 | 0 | 0 | |
| BjuA025267 | A07 | 1 | 1 | 3 | 0 | 0 | 0 | |
| BjuA042408 | A03 | 2 | 0 | 0 | 1 | 1 | 1 | |
| BjuB045293 | B05 | 0 | 0 | 0 | 0 | 0 | 0 | |
| P-1B-4 | AtHMA1 | 4 | 0 | 0 | 1 | 2 | 2 | 2 |
| BjuB002000 | B05 | 1 | 1 | 0 | 0 | 1 | 1 | |
| BjuB034828 | B02 | 0 | 0 | 0 | 0 | 0 | 0 | |
| BjuA036645 | A09 | 1 | 1 | 0 | 0 | 1 | 1 | |
表4 芥菜HMA家族基因启动子区顺式作用元件
Table 4 The cis-acting element in the promoter region of the HMA gene family in Brassica juncea
| 亚族 Subgroup | 基因 Gene | 染色体 Chr | 顺式作用元件数量 Number of cis-acting element | |||||
|---|---|---|---|---|---|---|---|---|
| G-Box | ABRE | LTR | TC | MBS | TGA | |||
| P-1B-1 | AtHMA5 | 1 | 1 | 1 | 0 | 2 | 0 | 2 |
| AtHMA6 | 4 | 0 | 1 | 0 | 1 | 1 | 1 | |
| AtHMA7 | 5 | 1 | 1 | 0 | 3 | 1 | 1 | |
| AtHMA8 | 5 | 0 | 1 | 0 | 1 | 2 | 1 | |
| BjuB028207 | B04 | 0 | 1 | 0 | 0 | 0 | 1 | |
| BjuA046861 | A09 | 0 | 0 | 0 | 0 | 0 | 0 | |
| BjuB019118 | B08 | 0 | 0 | 0 | 0 | 1 | 0 | |
| BjuB035323 | B02 | 0 | 0 | 0 | 1 | 0 | 0 | |
| BjuA038121 | A10 | 0 | 0 | 0 | 2 | 0 | 0 | |
| BjuB027969 | B04 | 0 | 0 | 1 | 3 | 1 | 0 | |
| BjuA033764 | A09 | 0 | 0 | 1 | 3 | 1 | 0 | |
| BjuA025022 | A06 | 1 | 1 | 0 | 0 | 1 | 0 | |
| BjuB036631 | B02 | 1 | 1 | 0 | 0 | 1 | 0 | |
| BjuA003262 | A01 | 0 | 0 | 1 | 0 | 0 | 0 | |
| BjuA038847 | A10 | 7 | 7 | 2 | 0 | 1 | 0 | |
| P-1B-2 | AtHMA2 | 4 | 1 | 0 | 0 | 1 | 2 | 0 |
| AtHMA3 | 4 | 1 | 0 | 1 | 0 | 2 | 2 | |
| AtHMA4 | 2 | 0 | 1 | 1 | 1 | 0 | 3 | |
| BjuA002855 | A01 | 0 | 0 | 0 | 0 | 1 | 0 | |
| BjuO011640 | Super_scaffold_184_753382_1581011 | 1 | 1 | 0 | 0 | 1 | 2 | |
| BjuA003595 | A01 | 0 | 0 | 0 | 0 | 0 | 0 | |
| BjuB040614 | B05 | 1 | 1 | 0 | 2 | 0 | 0 | |
| BjuA003596 | A01 | 2 | 2 | 2 | 1 | 0 | 3 | |
| BjuB040613 | B05 | 2 | 2 | 0 | 0 | 0 | 0 | |
| BjuA042404 | A03 | 2 | 3 | 2 | 0 | 1 | 0 | |
| BjuO003139 | Contig2023 | 2 | 2 | 1 | 0 | 0 | 0 | |
| BjuB035256 | B02 | 3 | 1 | 3 | 1 | 0 | 1 | |
| BjuA023298 | A06 | 1 | 1 | 0 | 2 | 0 | 0 | |
| BjuA025267 | A07 | 1 | 1 | 3 | 0 | 0 | 0 | |
| BjuA042408 | A03 | 2 | 0 | 0 | 1 | 1 | 1 | |
| BjuB045293 | B05 | 0 | 0 | 0 | 0 | 0 | 0 | |
| P-1B-4 | AtHMA1 | 4 | 0 | 0 | 1 | 2 | 2 | 2 |
| BjuB002000 | B05 | 1 | 1 | 0 | 0 | 1 | 1 | |
| BjuB034828 | B02 | 0 | 0 | 0 | 0 | 0 | 0 | |
| BjuA036645 | A09 | 1 | 1 | 0 | 0 | 1 | 1 | |
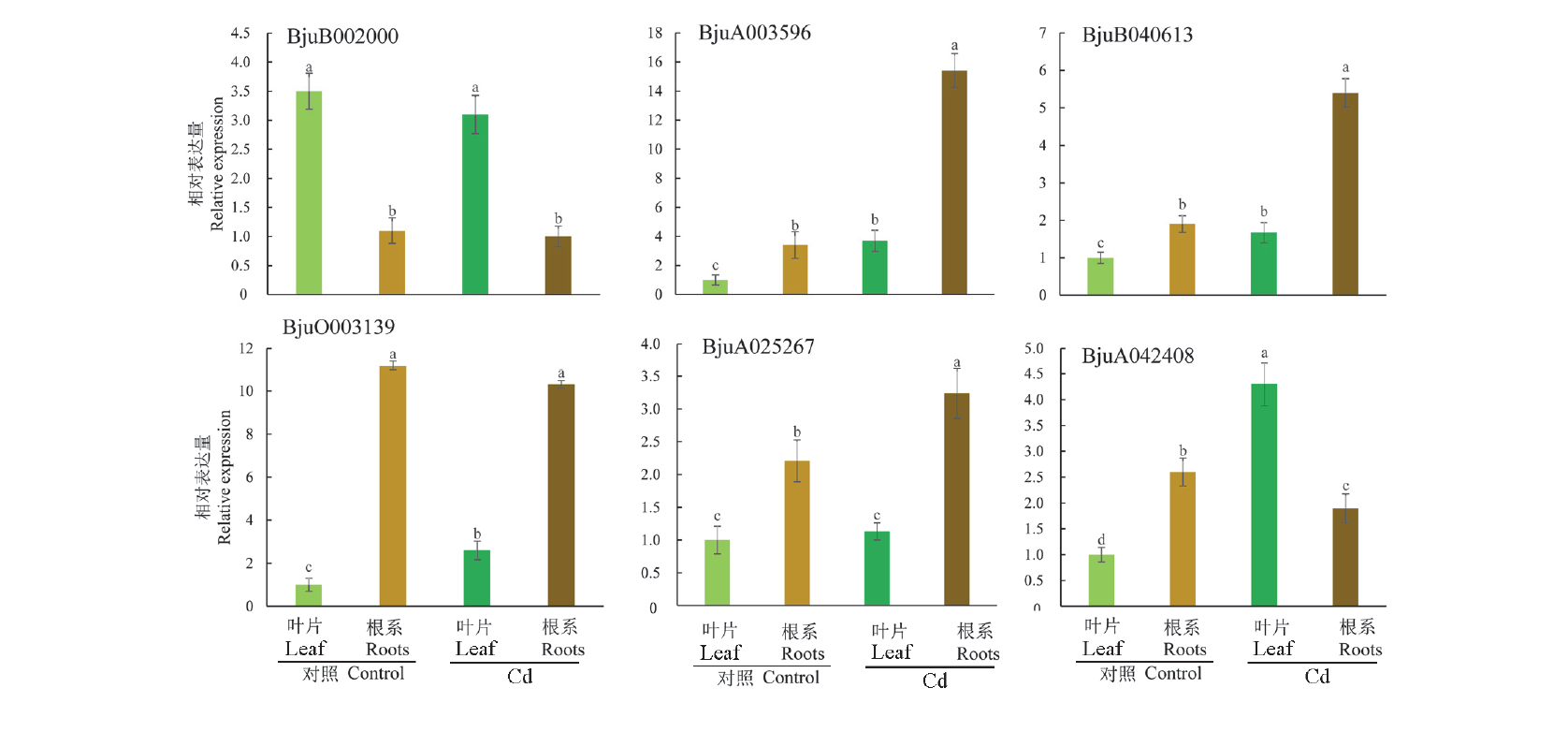
图5 芥菜HMA家族部分基因在不同部位及镉胁迫下表达水平的qRT-PCR分析 不同小写字母表示差异显著(P < 0.05)。
Fig. 5 Expression of HMA gene in Brassica juncea in different locations and under cadmium stress analyzed by qRT-PCR Different letters indicate significantly different values(P < 0.05).
| [1] | Arguëllo J M. 2003. Identification of ion-selectivity determinants in heavy-metal transport P1B-type ATPases. Molecular Membrane Biology, 195 (2):93-108. |
| [2] |
Axelsen K B, Palmgren M G. 2001. Inventory of the superfamily of P-type ion pumps in Arabidopsis. Plant Physiology, 126 (2):696-706.
pmid: 11402198 |
| [3] | Choudhury M R, Islam M S, Ahmed Z U, Nayar F. 2016. Phytoremediation of heavy metal contaminated buriganga riverbed sediment by Indian mustard and marigold plants. Environmental Progress & Sustainable Energy, 35 (1):117-124. |
| [4] |
Clemens S. 2001. Molecular mechanisms of plant metal tolerance and homeostasis. Planta, 212 (4):475-486.
doi: 10.1007/s004250000458 pmid: 11525504 |
| [5] |
Cola N A, Sancenon V, Navarro S R, Mayo S, Thiele D J, Ecker J R, Puig S, Penarrubia L. 2006. The Arabidopsis heavy metal P-type ATPase HMA 5 interacts with metallochaperones and functions in copper detoxification of roots. Plant Journal, 45 (2):225-236.
doi: 10.1111/tpj.2006.45.issue-2 URL |
| [6] |
Courbot M, Willems G, Motte P, Arvidsson S, Roosens N, Laprade P S, erbruggen N V. 2007. A major quantitative trait locus for cadmium tolerance in Arabidopsis halleri colocalizes with HMA4,a gene encoding a heavy metal ATPase. Plant Physiology, 144 (2):1052-1065.
doi: 10.1104/pp.106.095133 pmid: 17434989 |
| [7] |
Ghany S E A, Moule P M, Niyogi K K, Pilon M, Shikanai T. 2005. Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell, 17 (4):1233-1251.
doi: 10.1105/tpc.104.030452 URL |
| [8] |
Goswami S, Das S. 2015. A study on cadmium phytoremediation potential of indian mustard,Brassica juncea. International Journal of Phytoremediation, 17 (6):583-588.
doi: 10.1080/15226514.2014.935289 URL |
| [9] |
Gravot A, Lieutaud A, Verret F, Auroy P, Vavasseur A, Richaud P. 2004. AtHMA3,a plant P1B-ATPase,functions as a Cd/Pb transporter in yeast. Febs Letters, 561 (1-3):22-28.
doi: 10.1016/S0014-5793(04)00072-9 URL |
| [10] |
Hanikenne M, Talke I N, Haydon M J, Lanz C, Nolte A, Motte P, Kroymann J, Weigel D, Kramer U. 2008. Evolution of metal hyperaccumulation required cis-regulatory changes and triplication of HMA4. Nature, 453 (7193):391-395.
doi: 10.1038/nature06877 |
| [11] |
Hussain D, Haydon M J, Wang Y, Wong E, Sherson S M, Young J, Camakaris J, Harper J F, Cobbett C S. 2004. P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis. Plant Cell, 16 (5):1327-1339.
doi: 10.1105/tpc.020487 pmid: 15100400 |
| [12] |
Kim Y Y, Choi H, Segami S, Cho H T, Martinoia E, Masayoshi, Maeshima, Lee Y. 2009. AtHMA1 contributes to the detoxification of excess Zn(II) in Arabidopsis. Plant Journal, 58 (5):737-753.
doi: 10.1111/tpj.2009.58.issue-5 URL |
| [13] | Liao Tian-lu, Liao Tian-jiang, Lü Lin-hua. 2015. Application of phytoremediation technology in heavy metal contaminated soil. Beijing Agriculture, 6 (17):192. (in Chinese) |
| 廖天录, 廖天江, 吕麟华. 2015. 植物修复技术在重金属污染土壤中的应用. 北京农业, 6 (17):192. | |
| [14] |
Liu L, Yin H, Liu Y, Shen L, Yang X, Zhang D, Li M, Yan M. 2021. Analysis of cadmium-stress-induced microRNAs and their targets reveals bra-miR172b-3p as a potential Cd2+-specific resistance factor in Brassica juncea. Processes, 9 (7):1099.
doi: 10.3390/pr9071099 URL |
| [15] |
Miyadate H, Adachi S, Hiraizumi A, Tezuka K, Nakazawa N, Kawamoto T, Katou K, Kodama I, Sakurai K, Takahashi H, Nagasawa N S, Watanabe A, Fujimura T, Akagi H. 2011. OsHMA3,a P1B-type of ATPase affects root-to-shoot cadmium translocation in rice by mediating efflux into vacuoles. New Phytologist, 189 (1):190-199.
doi: 10.1111/nph.2010.189.issue-1 URL |
| [16] |
Morel M, Crouzet J, Gravot A, Auroy P, Leonhardt N, Vavasseur A, Richaud P. 2009. AtHMA3,a P1B-ATPase allowing Cd/Zn/Co/Pb vacuolar storage in Arabidopsis. Plant Physiology, 149 (2):894-904.
doi: 10.1104/pp.108.130294 URL |
| [17] |
Mourato M P, Moreira I N, Leitao I, Pinto F R, Sales J R, Martins L L. 2015. Effect of heavy metals in plants of the genus Brassica. International Journal of Molecular Sciences, 16 (8):17975-17998.
doi: 10.3390/ijms160817975 pmid: 26247945 |
| [18] |
Palmgren M G, Clemens S, Williams L E, Kramer U, Borg S, Schjorring J K, Sanders D. 2008. Zinc biofortification of cereals:problems and solutions. Trends In Plant Science, 13 (9):464-473.
doi: 10.1016/j.tplants.2008.06.005 pmid: 18701340 |
| [19] |
Papoyan A, Kochian L V. 2004. Identification of Thlaspi caerulescens genes that may be involved in heavy metal hyperaccumulation and tolerance Characterization of a novel heavy metal transporting ATPase. Plant Physiology, 136 (3):3814-3823.
pmid: 15516513 |
| [20] |
Parkin I A P, Gulden S M, Sharpe A G, Lukens L, Trick M, Osborn T C, Lydiate D J. 2005. Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics, 171 (2):765-781.
doi: 10.1534/genetics.105.042093 URL |
| [21] |
Puig S, Colas N A, Molina A G, Pearrubia L. 2007. Copper and iron homeostasis in Arabidopsis:responses to metal deficiencies,interactions and biotechnological applications. Plant Cell and Environment, 30 (3):271-290.
doi: 10.1111/j.1365-3040.2007.01642.x URL |
| [22] | Sankaran R P, Ebbs S D. 2008. Transport of Cd and Zn to seeds of Indian mustard(Brassica juncea)during specific stages of plant growth and development. Physiologia Plantarum, 132 (1):69-78. |
| [23] | Sun Tao, Zhang Yu-xiu, Chai Tuan-yao. 2011. Research progress on tolerance of Indian mustard(Brassica juncea L.) to heavy metal. Chinese Journal of Eco-Agriculture, 19 (1):226-234. (in Chinese) |
| 孙涛, 张玉秀, 柴团耀. 2011. 印度芥菜(Brassica juncea L.)重金属耐性机理研究进展. 中国生态农业学报, 19 (1):226-234. | |
| [24] |
Tan P, Zeng C, Wan C, Liu Z, Dong X, Peng J, Lin H, Li M, Liu Z, Yan M. 2021. Metabolic profiles of Brassica juncea roots in response to cadmium stress. Metabolites, 11 (6):383.
doi: 10.3390/metabo11060383 URL |
| [25] |
Verret F, Gravot A, Auroy P, Preveral S, Forestier C, Vavasseur A, Richaud P. 2005. Heavy metal transport by AtHMA4 involves the N-terminal degenerated metal binding domain and the C-terminal His11 stretch. Febs Letters, 579 (6):1515-1522.
doi: 10.1016/j.febslet.2005.01.065 URL |
| [26] |
Verreta F, Gravota A, Auroya P, Leonhardta N, Davidb P, Nussaumeb L, Vavasseura A, Richaud P. 2004. Overexpression of AtHMA4enhances root-to-shoot translocation of zinc and cadmium and plant metal tolerance. Febs Letters, 576 (3):306-312.
doi: 10.1016/j.febslet.2004.09.023 URL |
| [27] |
Wang Ruonan, Nie Lanchun, Zhang Shuangshuang, Cui Qiang, Jia Mingfei. 2019. Research progress on plant resistance to heavy metal stress. Acta Horticulturae Sinica, 46 (1):157-170. (in Chinese)
doi: 10.16420/j.issn.0513-353x.2018-0585 URL |
|
王若男, 乜兰春, 张双双, 崔强, 贾明飞. 2019. 植物抗重金属胁迫研究进展. 园艺学报, 46 (1):157-170.
doi: 10.16420/j.issn.0513-353x.2018-0585 URL |
|
| [28] | Wang Xiao-tong, Li Hao-yang, Xu Ji-chen. 2014. Bioinformatics analysis of the heavy metal transporting ATPase gene family in poplar genome. Plant Physiology Journal, 50 (7):891-900. (in Chinese) |
| 王晓桐, 李昊阳, 徐吉臣. 2014. 毛果杨HMA基因家族的生物信息学分析. 植物生理学报, 50 (7):891-900. | |
| [29] |
Williams L E, Mills R F. 2005. P(1B)-ATPases--an ancient family of transition metal pumps with diverse functions in plants. Trends In Plant Science, 10 (10):491-502.
doi: 10.1016/j.tplants.2005.08.008 pmid: 16154798 |
| [30] |
Wong C K E, Cobbett C S. 2009. HMA P-type ATPases are the major mechanism for root-to-shoot Cd translocation in Arabidopsis thaliana. New Phytologist, 181 (1):71-78.
doi: 10.1111/nph.2009.181.issue-1 URL |
| [31] |
Wong C K E, Jarvis R S, Sherson S M, Cobbett C S. 2009. Functional analysis of the heavy metal binding domains of the Zn/Cd-transporting ATPase,HMA2,in Arabidopsis thaliana. New Phytologist, 181 (1):79-88.
doi: 10.1111/nph.2009.181.issue-1 URL |
| [32] |
Xu D, Lu Z C, Jin K M, Qiu W M, Qiao G R, Han X J, Zhuo R Y. 2020. SPDE:a multi-functional software for sequence processing and data extraction. Bioinformatics, 37 (20):3686-3687.
doi: 10.1093/bioinformatics/btab235 URL |
| [33] | Zhang Fu-gui, Xiao Xin, Yan Gui-xin. 2017. Evolution of HMA gene family in Brassica AC genomes. Chinese Journal of Oil Crop Sciences, 39 (3):294-307. (in Chinese) |
| 张付贵, 肖欣, 闫贵欣. 2017. HMA基因家族在芸薹属AC基因组中的进化分析. 中国油料作物学报, 39 (3):294-307. | |
| [34] | Zhang Yu-xiu, Chai Tuan-yao. 2006. Isolation and function of heavy metal responsive gene in plant. Beijing: China Agriculture Press. (in Chinese) |
| 张玉秀, 柴团耀. 2006. 植物重金属调节基因的分离和功能. 北京: 中国农业出版社. |
| [1] | 高鹏飞, 高冰, 冯郑红, 吴建慧. 绢毛委陵菜PsWRKY40的克隆与耐镉功能分析[J]. 园艺学报, 2023, 50(6): 1269-1283. |
| [2] | 张琨, 思彬彬, 周军, 任玉锋, 张欣, 徐文娣, 王佳伟, 乔帅, 王惠冉. 苹果砧木‘青砧1号’叶片cDNA文库构建及MdMLO上游调控因子的筛选[J]. 园艺学报, 2023, 50(5): 933-946. |
| [3] | 陈道宗, 刘镒, 沈文杰, 朱博, 谭晨. 白菜、甘蓝和甘蓝型油菜PAP1/2同源基因的鉴定及分析[J]. 园艺学报, 2022, 49(6): 1301-1312. |
| [4] | 杨天宸, 陈晓童, 吕可, 张荻. 百子莲脱水素基因ApSK3对逆境与激素信号的应答模式与调控机制[J]. 园艺学报, 2021, 48(8): 1565-1578. |
| [5] | 孟秋峰*,王毓洪,黄芸萍,王 洁,任锡亮,高天一. 茎瘤芥新品种‘甬榨4号’[J]. 园艺学报, 2020, 47(6): 1219-1220. |
| [6] | 张梦原1,2,谢幸男1,李 盈1,葛晓霞1,*. 柑橘CsHB1启动子的克隆及功能分析[J]. 园艺学报, 2018, 45(8): 1491-1500. |
| [7] | 李朝闯1,马关鹏1,2,谢 婷1,3,陈 娇1,王志敏1,宋 明1,汤青林1,*. 芥菜AGL18家族成员与开花整合子SOC1的互作分析[J]. 园艺学报, 2017, 44(3): 463-474. |
| [8] | 李朝闯,马关鹏,杨修勤,王志敏,宋 明,汤青林. 芥菜开花抑制因子SVP表达分析及其与FLC互作的调节位点鉴定[J]. 园艺学报, 2016, 43(8): 1513-1524. |
| [9] | 孟秋峰1,汪炳良2,*,王毓洪1,任锡亮1,黄芸萍1. 春茎瘤芥新品种‘甬榨5号’[J]. 园艺学报, 2016, 43(4): 809-810. |
| [10] | 曹明明,杨 佳,李晓屿,李玉花,蓝兴国. 羽衣甘蓝ARC1与Exo70A1蛋白相互作用结构域的分析与鉴定[J]. 园艺学报, 2015, 42(4): 791-798. |
| [11] | 谢 婷,谷慧英,江 为,马关鹏,陈 娇,王志敏,宋 明,汤青林. 芥菜开花整合子SOC1与AGL24蛋白K结构域的互作位点鉴定[J]. 园艺学报, 2015, 42(3): 554-562. |
| [12] | 邓 帅1,*,成妮妮2,*,丁瑞瑞1,刘 宇1,张元湖1,**. 苹果和梨AFS基因启动子的克隆、序列比对及功能分析[J]. 园艺学报, 2015, 42(12): 2353-2361. |
| [13] | 陈 娇*,赵夏云*,鲜登宇*,马关鹏,谢 婷,王志敏,宋 明**,汤青林**. 芥菜开花整合子SOC1启动子的克隆及其与FLC、SVP蛋白互作的研究[J]. 园艺学报, 2015, 42(10): 1931-1943. |
| [14] | 孙 君 陈桂信 叶乃兴 吕恃衡 刘志钦 黄 玮 林志达. 茉莉花香气相关基因JsDXS及其启动子的克隆与表达分析[J]. 园艺学报, 2014, 41(6): 1236-1244. |
| [15] | 符聪慧, 王建平, 张冲, 马锋旺, 张军科. 苹果PGIP基因部分家族成员启动子的克隆与功能分析[J]. 园艺学报, 2014, 41(11): 2169-2178. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司