
园艺学报 ›› 2023, Vol. 50 ›› Issue (3): 534-548.doi: 10.16420/j.issn.0513-353x.2022-0003
毛可欣1,2, 安淼1, 王海荣1, 王世金3, 吕巍2, 郭盈添1, 李健1,*( ), 李国田1,*(
), 李国田1,*( )
)
收稿日期:2022-09-03
修回日期:2022-12-30
出版日期:2023-03-25
发布日期:2023-04-03
通讯作者:
*(E-mail:lijian097597@163.com,ligt2008@163.com)
基金资助:
MAO Kexin1,2, AN Miao1, WANG Hairong1, WANG Shijin3, LÜ Wei2, GUO Yingtian1, LI Jian1,*( ), LI Guotian1,*(
), LI Guotian1,*( )
)
Received:2022-09-03
Revised:2022-12-30
Online:2023-03-25
Published:2023-04-03
Contact:
*(E-mail:lijian097597@163.com,ligt2008@163.com)
摘要:
通过对‘红阳’猕猴桃基因组鉴定得到277个MYB家族成员并将其分为12个亚家族(S1 ~ S12),其中多数序列属于R2R3-MYB,其次是MYB相关蛋白,仅鉴定到1个4R-MYB。S11和S12亚家族在进化上处于较早的位置,结构较为多样化,而S1和S2在进化上出现较晚。顺式作用元件分析显示,该家族主要含水杨酸、赤霉素和茉莉酸甲酯等相关植物激素元件,还有部分参与昼夜节律、低温、细胞周期等相关元件。密码子偏好性分析显示,猕猴桃MYB家族有3个高频密码子(UUG、AGA和AGG),基因的密码子偏好使用A/T,第3位碱基偏好更强。通过顺式作用元件分析和4个猕猴桃品种越冬期转录组数据分析筛选了9个可能参与低温胁迫响应MYB基因,对其在低温胁迫下的软枣猕猴桃‘龙成2号’中进行检测验证,确认这些基因参与猕猴桃对低温的响应。
中图分类号:
毛可欣, 安淼, 王海荣, 王世金, 吕巍, 郭盈添, 李健, 李国田. 猕猴桃MYB家族成员鉴定及其低温表达分析[J]. 园艺学报, 2023, 50(3): 534-548.
MAO Kexin, AN Miao, WANG Hairong, WANG Shijin, LÜ Wei, GUO Yingtian, LI Jian, LI Guotian. Identification and Low Temperature Expression Analysis of MYB Transcription Factor Family in Kiwifruit[J]. Acta Horticulturae Sinica, 2023, 50(3): 534-548.
| 基因Gene | 上游引物(5′-3′)Forward primer | 下游引物(5′-3′)Reverse primer |
|---|---|---|
| AcMYB200 | GAGATACCAGTGGCGGAGAG | CCCTGTATGCCGTTGCTTAT |
| AcMYB61 | TCTTGGATGGTGTTGTTCCA | CCTGATCTGCTACGCTTTCC |
| AcMYB63 | GAGGTGCTTAAGCCAACAGC | GTCCGACTTCCAAACTGCAT |
| AcMYB56 | GTTCATAAGAGCGGGACCAA | GGAAGCATTTGGGTCAAAAA |
| AcMYB71 | GAACCGGTGGTATTGAAACG | ACACGATCTCCACCTTTTGG |
| AcMYB123 | CGACAAGTTCCTCGAAGCTC | TTTTGCGGGTAAGGATGAAC |
| AcMYB210 | TGTGAATCCTCCACAAGCTG | ATGTGTCCCCGTTTTAGCAG |
| AcMYB235 | CGACCTGAGCAAGAAGATCC | CTTCTGTGCATGGCTTCGTA |
| Actin | GTGCTCAGTGGTGGTTCAA | GACGCTGTATTTCCTCTCAG |
表1 本研究中的引物序列
Table 1 Primers in this study
| 基因Gene | 上游引物(5′-3′)Forward primer | 下游引物(5′-3′)Reverse primer |
|---|---|---|
| AcMYB200 | GAGATACCAGTGGCGGAGAG | CCCTGTATGCCGTTGCTTAT |
| AcMYB61 | TCTTGGATGGTGTTGTTCCA | CCTGATCTGCTACGCTTTCC |
| AcMYB63 | GAGGTGCTTAAGCCAACAGC | GTCCGACTTCCAAACTGCAT |
| AcMYB56 | GTTCATAAGAGCGGGACCAA | GGAAGCATTTGGGTCAAAAA |
| AcMYB71 | GAACCGGTGGTATTGAAACG | ACACGATCTCCACCTTTTGG |
| AcMYB123 | CGACAAGTTCCTCGAAGCTC | TTTTGCGGGTAAGGATGAAC |
| AcMYB210 | TGTGAATCCTCCACAAGCTG | ATGTGTCCCCGTTTTAGCAG |
| AcMYB235 | CGACCTGAGCAAGAAGATCC | CTTCTGTGCATGGCTTCGTA |
| Actin | GTGCTCAGTGGTGGTTCAA | GACGCTGTATTTCCTCTCAG |
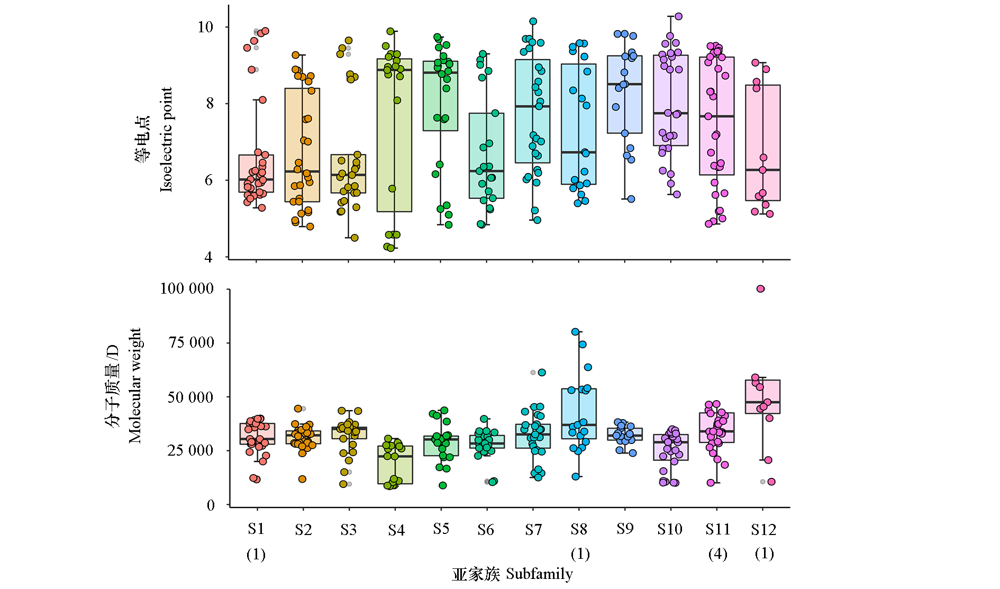
图2 猕猴桃MYB家族蛋白的等电点和分子质量 括号内数字表示分子量超过100 000 D的MYB蛋白数量。
Fig. 2 Protein isoelectric point and molecular weight of kiwifruit MYB family The number in parenthes indicates the number of MYB proteins which molecular weight exceeding 100 000 D.

图3 猕猴桃MYB家族的基因保守基序motif(A)、蛋白保守结构域(B)和基因结构(C)分布
Fig. 3 Distribution of conserved motif(A),protein conserved domain(B)and genetic structure(C)of kiwifruit MYB gene family
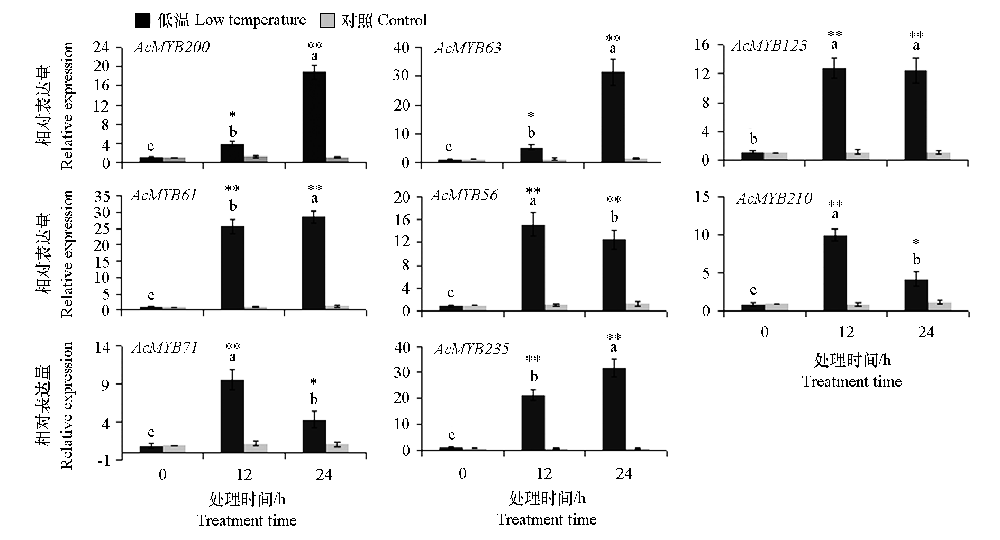
图8 低温处理下猕猴桃8个MYB基因的表达 不同字母表示不同处理时间在0.05水平上差异显著。*、**表示处理与对照在0.05和0.01水平差异显著。
Fig. 8 Expression analysis of eight kiwifruit MYB genes under low temperature treatment Different letters indicate that there are significant differences in genes at the level of 0.05 under different treatment times. *,** indicated significant difference between treatment and control at 0.05 and 0.01 levels,respectively.
| [1] |
Aasland R, Stewart A F, Gibson T. 1996. The SANT domain:a putative DNA-binding domain in the SWI-SNF and ADA complexes,the transcriptional co-repressor N-CoR and TFIIIB. Trends in Biochemical Sciences, 21:87-88.
pmid: 8882580 |
| [2] |
Anwar M, Yu W, Yao H, Zhou P, Allan A C, Zeng L. 2019. NtMYB3,an R2R3-MYB from narcissus,regulates flavonoid biosynthesis. International Journal of Molecular Sciences, 20:5456.
doi: 10.3390/ijms20215456 URL |
| [3] | Ashesh N. 2002. Investigations on evolutionary changes in base distributions in gene sequences. Internet Electronic Journal of Molecular Design, 1 (10):545. |
| [4] |
Baldoni E, Genga A, Cominelli E. 2015. Plant MYB transcription factors:their role in drought response mechanisms. International Journal of Molecular Sciences, 16:15811-15851.
doi: 10.3390/ijms160715811 pmid: 26184177 |
| [5] |
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden T L. 2009. BLAST+:architecture and applications. BMC Bioinformatics, 10 (421):1-9.
doi: 10.1186/1471-2105-10-1 |
| [6] |
Cao Y, Li K, Li Y, Zhao X, Wang L. 2020. MYB transcription factors as regulators of secondary metabolism in plants. Biology, 9 (3):61-76.
doi: 10.3390/biology9030061 URL |
| [7] |
Chen C, Chen H, Zhang Y, Thomas H R, Frank M H, He Y, Xia R. 2020. TBtools:an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 13 (8):1194-1202.
doi: 10.1016/j.molp.2020.06.009 URL |
| [8] |
Chen Y H, Wu X M, Ling H Q, Yang W C. 2006a. Transgenic expression of DwMYB2 impairs iron transport from root to shoot in Arabidopsis thaliana. Cell Research, 16:830-840.
doi: 10.1038/sj.cr.7310099 |
| [9] |
Chen Y H, Yang X Y, He K, Liu M H, Li J G, Luo J C, Deng X W, Chen Z L, Gu H Y, Qu L J. 2006b. The MYB transcription factor superfamily of Arabidopsis:expression analysis and phylogenetic comparison with the rice MYB family. Plant Molecular Biology, 60:107-124.
doi: 10.1007/s11103-005-2910-y URL |
| [10] | Das G, Duncan L R. 2014. Configuration of wobble base pairs having pyrimidines as anticodon wobble bases: significance for codon degeneracy. Journal of Biomolecular Structure and Dynamics, 329:1500-1520. |
| [11] |
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. 2010. MYB transcription factors in Arabidopsis. Trends in Plant Science, 15:573-581.
doi: 10.1016/j.tplants.2010.06.005 URL |
| [12] |
Enany S. 2014. Structural and functional analysis of hypothetical and conserved proteins of Clostridium tetani. Journal of Infection and Public Health, 7 (4):296-307.
doi: 10.1016/j.jiph.2014.02.002 URL |
| [13] |
Feng G, Burleigh J G, Braun E L, Mei W, Barbazuk W B. 2017. Evolution of the 3R-MYB gene family in plants. Genome Biology and Evolution, 9:1013-1029.
doi: 10.1093/gbe/evx056 pmid: 28444194 |
| [14] | Gupta S K, Majumdar S K, Bhattacharya T. 2000. Studies on the relationships between the synonymous codon usage and protein secondary structural units. Biochemical & Biophysical Research Communications, 269 (3):692-696. |
| [15] |
Haupt S, Ziegler A, Cowan G, Torrance L. 2009. Studies of the role and function of barley stripe mosaic virus encoded proteins in replication and movement using GFP fusions. Methods in Molecular Biology, 515:287-297.
doi: 10.1007/978-1-59745-559-6_20 pmid: 19378124 |
| [16] | Hou Qian-qian. 2019. Expression strategy and application of exogenous genes in Candida glycerinogenes[Ph. D. Dissertation]. Wuxi: Jiangnan University. (in Chinese) |
| 侯倩倩. 2019. 外源基因在产甘油假丝酵母中的表达策略及应用研究[博士论文]. 无锡: 江南大学. | |
| [17] | Huang S X, Ding J, Deng D J, Tang W, Sun H H, Xiao F M, Wang H L, Zheng H K, Fei Z J, Liu Y S. 2013. Draft genome of the kiwifruit Actinidia chinensis. Nature Communications, 4 (4):26-40. |
| [18] | Hughes A L. 1994. The evolution of functionally novel proteins after gene duplication. Proceedings of the Royal Society B-biological Sciences, 256:119-124. |
| [19] |
Jia J, Xue Q Z. 2009. Codon usage biases of transposable elements and host nuclear genes in Arabidopsis thaliana and Oryza sativa. Genomics Proteomics Bioinformatics, 7 (4):175-184.
doi: 10.1016/S1672-0229(08)60047-9 URL |
| [20] |
Jiang C K, Rao G Y. 2020. Insights into the diversification and evolution of R2R3-MYB transcription factors in plants. Plant Physiology, 183:637-655.
doi: 10.1104/pp.19.01082 URL |
| [21] |
Kanei-Ishii C, Sarai A, Sawazaki T, Nakagoshi H, He D N. 1990. The tryptophan cluster:a hypothetical structure of the DNA-binding domain of the MYB protooncogene product. Journal of Biological Chemistry, 265:19990-19995.
pmid: 2246275 |
| [22] | Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y. 2002. PlantCARE,a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research, 30:325-327. |
| [23] |
Letunic I, Khedkar S, Bork P. 2020. SMART:recent updates,new developments and status in 2020. Nucleic Acids Research, 49:458-460.
doi: 10.1093/nar/gkaa1189 URL |
| [24] |
Li Guisheng. 2021. Comparative analysis of‘Jinyan’and‘Hongyang’kiwifruit transcriptomes. Acta Horticulturae Sinica, 48 (6):1183-1196. (in Chinese)
doi: 10.16420/j.issn.0513-353x.2020-0510 |
|
李贵生. 2021. 猕猴桃‘金艳’和‘红阳’果实转录组的比较分析. 园艺学报, 48 (6):1183-1196.
doi: 10.16420/j.issn.0513-353x.2020-0510 |
|
| [25] | Li J, Han G, Sun C, Sui N. 2019. Research advances of MYB transcription factors in plant stress resistance and breeding. Plant Signaling & Behavior, 14 (8):e1613131. |
| [26] |
Li W, Ding Z, Ruan M, Yu X, Peng M, Liu Y. 2017. Kiwifruit R2R3-MYB transcription factors and contribution of the novel AcMYB75 to red kiwifruit anthocyanin biosynthesis. Scientific Reports, 7 (1):16861.
doi: 10.1038/s41598-017-16905-1 |
| [27] |
Liu C, Xie T, Chen C, Luan A, Long J, Li C, Ding Y, Yehua H. 2017. Genome-wide organization and expression profiling of the R2R3-MYB transcription factor family in pineapple(Ananas comosus). BMC Genomics, 18:503-518.
doi: 10.1186/s12864-017-3896-y URL |
| [28] |
Liu J, Osbourn A, Ma P. 2015. MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Molecular Plant, 8:689-708.
doi: 10.1016/j.molp.2015.03.012 pmid: 25840349 |
| [29] | Lu S, Wang J, Chitsaz F, Derbyshire M K, Geer R C. 2020. CDD/SPARCLE:the conserved domain database in 2020. Nucleic Acids Research, 48:265-268. |
| [30] | Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar Gustavo A. 2020. Pfam:the protein families database in 2021. Nucleic Acids Research, 49:412-419. |
| [31] |
Novembre J A. 2002. Accounting for background nucleotide composition when measuring codon usage bias. Molecular Biology and Evolution, 19 (8):1390-1394.
pmid: 12140252 |
| [32] |
Potter S C, Luciani A, Eddy S R, Park Y, Lopez R, Finn R D. 2018. HMMER web server:2018 update. Nucleic Acids Research, 46:200-204.
doi: 10.1093/nar/gky448 pmid: 29905871 |
| [33] | Roy S. 2016. Function of MYB domain transcription factors in abiotic stress and epigenetic control of stress response in plant genome. Plant Signaling & Behavior, 11:e1117723. |
| [34] | Shi Yang, Xu Xiao, Li Hao-yang, Xu Qian, Xu Jichen. 2014. Bioinformatics analysis of the expansin gene family in rice. Hereditas, 36 (8):809-820. (in Chinese) |
| 施杨, 徐筱, 李昊阳, 徐倩, 徐吉臣. 2014. 水稻扩展蛋白家族的生物信息学分析. 遗传, 36 (8):809-820. | |
| [35] |
Sueoka N. 2001. Near homogeneity of PR2-bias fingerprints in the human genome and their implications in phylogenetic analyses. Journal of Molecular Evolution, 53 (4-5):469-476.
pmid: 11675607 |
| [36] | Suzuki Y. 2011. Statistical methods for detecting natural selection from genomic data. Genes & Genetic Systems, 856:359-376. |
| [37] |
Tamura K, Stecher G, Kumar S. 2021. MEGA11:molecular evolutionary genetics analysis version 11. Molecular Biology and Evolution, 38:3022-3027.
doi: 10.1093/molbev/msab120 URL |
| [38] | Tian F, Yang D C, Meng Y Q, Jin J, Gao G. 2020. PlantRegMap:charting functional regulatory maps in plants. Nucleic Acids Research, 48:1104-1113. |
| [39] |
Wang L, Roossinck M J. 2006. Comparative analysis of expressed sequences reveals a conserved pattern of optimal codon usage in plants. Plant Molecular Biology, 61:699-710.
doi: 10.1007/s11103-006-0041-8 pmid: 16897485 |
| [40] | Wang L, Tang W, Hu Y, Zhang Y, Sun J. 2019. A MYB/bHLH complex regulates tissue-specific anthocyanin biosynthesis in the inner pericarp of red-centered kiwifruit Actinidia chinensis cv. Hongyang. Plant Journal, 99:359-378. |
| [41] |
Wang X, Niu Y, Zheng Y. 2021. Multiple functions of MYB transcription factors in abiotic stress responses. International Journal of Molecular Science, 22:6125.
doi: 10.3390/ijms22116125 URL |
| [42] |
Wang Y, Tang H, Debarry J D, Tan X, Li J. 2012. MCScanX:a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Research, 40:e49.
doi: 10.1093/nar/gkr1293 URL |
| [43] |
Wilkins M R, Gasteiger E, Bairoch A, Sanchez J C, Williams K L. 1999. Protein identification and analysis tools in the ExPASy server. Methods in Molecular Biology, 112:531-552.
pmid: 10027275 |
| [44] |
Wu H L, Ma T, Kang M H, Ai F D, Zhang J L, Dong G Y, Liu J Q. 2019. A high-quality Actinidia chinensis(kiwifruit)genome. Horticulture Research, 6:117.
doi: 10.1038/s41438-019-0202-y |
| [45] |
Wu Jiacheng, Wang Guiqing, Muhammad Anwar, Zeng Lihui. 2018. Cloning and functional analysis of R2R3-MYB gene NtMYB5 in Narcissus tazetta var. chinensis. Acta Horticulturae Sinica, 45 (7):1327-1337. (in Chinese)
doi: 10.16420/j.issn.0513-353x.2017-0889 |
|
吴嘉诚, 王桂青, Muhammad Anwar, 曾黎辉. 2018. 中国水仙R2R3-MYB 基因NtMYB5的克隆和功能研究. 园艺学报, 45 (7):1327-1337.
doi: 10.16420/j.issn.0513-353x.2017-0889 |
|
| [46] |
Yan H L, Pei X N, Zhang H, Li X, Zhang X X, Zhao M H, Chiang V L, Sederoff R R, Zhao X Y. 2021. MYB-mediated regulation of anthocyanin biosynthesis. International Journal of Molecular Sciences, 22 (6):3103-3128.
doi: 10.3390/ijms22063103 URL |
| [47] |
Yan X W, Hoek T A, Vale R D, Marvin M E. 2016. Dynamics of translation of single mRNA molecules invivo. Cell, 165:976-989.
doi: 10.1016/j.cell.2016.04.034 URL |
| [48] | Yang Ping. 2011. Characteristics analysis of codon usage of rice HKT gene family. Journal of Shanxi Agricultural Sciences, 39 (11):1137-1140. (in Chinese) |
| 杨平. 2011. 水稻 HKT 基因家族密码子使用特性分析. 山西农业科学, 39 (11):1137-1140. | |
| [49] |
Yao G F, Ming M L, Allan A C, Gu C, Li L T, Wu X, Wang R Z, Chang Y J, Qi K J, Zhang S L, Wu J. 2017. Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant Journal, 92:437-451.
doi: 10.1111/tpj.2017.92.issue-3 URL |
| [50] |
Yu M, Man Y, Wang Y. 2019. Light-and temperature-induced expression of an R2R3-MYB gene regulates anthocyanin biosynthesis in red-fleshed kiwifruit. International Journal of Molecular Sciences, 20 (20):5228-5249.
doi: 10.3390/ijms20205228 URL |
| [51] |
Yue J Y, Liu J C, Tang W, Wu Y Q, Tang X F, Li W, Yang Y, Wang L H, Huang S X, Fang C B, Zhao K, Fei Z J, Liu Y S, Zheng Y. 2020. Kiwifruit Genome Database(KGD):a comprehensive resource for kiwifruit genomics. Horticulture Research, 7 (1):117-124.
doi: 10.1038/s41438-020-0338-9 |
| [52] | Zhang H, Li J, Wang R X, Zhi J K, Yin P, Xu J C. 2019. Comparative analysis of codon usage pattern of expansin genes from eight plant species. Journal of Biomolecular Structure & Dynamics, 37 (4):910-917. |
| [1] | 俞沁含, 李俊铎, 崔莹, 王佳慧, 郑巧玲, 徐伟荣. 山葡萄转录因子VaMYB4a互作蛋白的筛选与鉴定[J]. 园艺学报, 2023, 50(3): 508-522. |
| [2] | 王泉城, 武军, 李磊, 石延霞, 谢学文, 李宝聚, 柴阿丽. 多主棒孢菌CcTLS1对黄瓜的致病机理分析[J]. 园艺学报, 2023, 50(3): 569-582. |
| [3] | 王泽涵, 于文涛, 王鹏杰, 刘财国, 樊晓静, 谷梦雅, 蔡春平, 王攀, 叶乃兴. 茶树秃房与茸房种质花器官差异表达基因的WGCNA分析[J]. 园艺学报, 2023, 50(3): 620-634. |
| [4] | 王晓晨, 聂子页, 刘先菊, 段伟, 范培格, 梁振昌. 脱落酸对‘京香玉’葡萄果实单萜物质合成的影响[J]. 园艺学报, 2023, 50(2): 237-249. |
| [5] | 任菲, 卢苗苗, 刘吉祥, 陈信立, 刘道凤, 眭顺照, 马婧. 蜡梅胚胎晚期丰富蛋白基因CpLEA的表达及抗性分析[J]. 园艺学报, 2023, 50(2): 359-370. |
| [6] | 翟含含, 翟宇杰, 田义, 张叶, 杨丽, 温陟良, 陈海江. 桃SAUR家族基因分析及PpSAUR5功能鉴定[J]. 园艺学报, 2023, 50(1): 1-14. |
| [7] | 袁馨, 徐云鹤, 张雨培, 单楠, 陈楚英, 万春鹏, 开文斌, 翟夏琬, 陈金印, 甘增宇. 猕猴桃后熟过程中ABA响应结合因子AcAREB1调控AcGH3.1的表达[J]. 园艺学报, 2023, 50(1): 53-64. |
| [8] | 蔺海娇, 梁雨晨, 李玲, 马军, 张璐, 兰振颖, 苑泽宁. 薰衣草CBF途径相关耐寒基因挖掘与调控网络分析[J]. 园艺学报, 2023, 50(1): 131-144. |
| [9] | 赵雪艳, 王琪, 王莉, 王方圆, 王庆, 李艳. 基于比较转录组的延胡索组织差异性表达分析[J]. 园艺学报, 2023, 50(1): 177-187. |
| [10] | 宋 放, 陈 奇, 袁炎良, 陈 沙, 尹海军, 蒋迎春, . 黄肉猕猴桃新品种‘先沃1号’[J]. 园艺学报, 2022, 49(S2): 47-48. |
| [11] | 齐永杰, 高正辉, 马 娜, 王清明, 柯凡君, 陈 钱, 徐义流, . 黄肉抗溃疡病猕猴桃新品种‘皖农金果’[J]. 园艺学报, 2022, 49(S2): 49-50. |
| [12] | 张慧琴, 楼国荣, 陆玲鸿, 古咸彬, 宋根华, 谢 鸣. 黄肉中华猕猴桃新品种‘金义’[J]. 园艺学报, 2022, 49(S2): 51-52. |
| [13] | 高彦龙, 吴玉霞, 张仲兴, 王双成, 张瑞, 张德, 王延秀. 苹果ELO家族基因鉴定及其在低温胁迫下的表达分析[J]. 园艺学报, 2022, 49(8): 1621-1636. |
| [14] | 邱子文, 刘林敏, 林永盛, 林晓洁, 李永裕, 吴少华, 杨超. 千层金MbEGS基因的克隆与功能分析[J]. 园艺学报, 2022, 49(8): 1747-1760. |
| [15] | 陶鑫, 朱荣香, 贡鑫, 吴磊, 张绍铃, 赵建荣, 张虎平. 梨果糖激酶基因PpyFRK5在果实蔗糖积累中的作用[J]. 园艺学报, 2022, 49(7): 1429-1440. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司