
园艺学报 ›› 2023, Vol. 50 ›› Issue (2): 382-396.doi: 10.16420/j.issn.0513-353x.2021-1114
田明康1, 徐智祥1, 刘秀群2, 眭顺照1, 李名扬1, 李志能1,*( )
)
收稿日期:2022-05-19
修回日期:2022-11-05
出版日期:2023-02-25
发布日期:2023-03-06
通讯作者:
*(E-mail:znli@swu.edu.cn)
基金资助:
TIAN Mingkang1, XU Zhixiang1, LIU Xiuqun2, SUI Shunzhao1, LI Mingyang1, LI Zhineng1,*( )
)
Received:2022-05-19
Revised:2022-11-05
Online:2023-02-25
Published:2023-03-06
Contact:
*(E-mail:znli@swu.edu.cn)
摘要:
对蜡梅(Chimonanthus praecox)AP2(Apetala2,AP2)亚家族成员进行全基因组鉴定,并分析其在花发育过程中的表达模式;研究CpAP2-L11的表达特异性,过表达拟南芥(Arabidopsis thaliana)并观察其表型。共鉴定出20个蜡梅AP2亚家族转录因子,其中euAP2、basalANT和euANT组分别有5、6和9个成员,且euAP2组全部成员无miR172结合位点。共线性分析发现有12个成员形成了16对复制基因,并在进化过程中受纯化选择。多数AP2亚家族成员在4、5月花芽中高表达,在花芽进行需冷量积累的过程或后期低表达,仅CpAP2-L11在花芽需冷量积累到570 CU(Chill units,CU)的始花期高表达,同时CpAP2-L11在蜡梅幼果、外轮花被片和雄蕊中高表达,在茎、叶中低表达,并受高温和低温诱导表达量降低。拟南芥异源表达CpAP2-L11,抽薹时间显著提前,且FT、SOC1、LFY和AP1基因表达量显著升高。蜡梅CpAP2-L11可能参与低温诱导打破蜡梅休眠导致寒冬开花及春季花芽分化,且促进过表达拟南芥早开花。
中图分类号:
田明康, 徐智祥, 刘秀群, 眭顺照, 李名扬, 李志能. 蜡梅AP2亚家族转录因子鉴定及CpAP2-L11功能研究[J]. 园艺学报, 2023, 50(2): 382-396.
TIAN Mingkang, XU Zhixiang, LIU Xiuqun, SUI Shunzhao, LI Mingyang, LI Zhineng. Identification of the AP2 Subfamily Transcription Factors in Chimonanthus praecox and the Functional Study of CpAP2-L11 [J]. Acta Horticulturae Sinica, 2023, 50(2): 382-396.
| 用途 Usage | 引物名称 Primer name | 序列(5′-3′) Sequence |
|---|---|---|
| CpAP2-L11基因克隆引物 CpAP2-L11 gene cloning primer | CpAP2-L11-F | AACAAATCGGACGGTTCAAACCACT |
| CpAP2-L11-R | GAAAATCAAAAACCGATCTCAGCCGG | |
| 表达载体构建引物 Expression vector construction primer | eCpAP2-L11-F | CGGGGTACCATGGGGAAACCATCACAGAAG |
| eCpAP2-L11-R | ACGCGTCGACTTACTGTCCATCCCAGTCGGC | |
| 实时荧光定量PCR qRT-PCR primer | qCpAP2-L11-F | TCCCGACCTTAACGTCTCGTCAGAG |
| qCpAP2-L11-R | GCAGCGGGGAGTTTTGTCGGATTCT | |
| 蜡梅内参基因实时荧光定量PCR qRT-PCR primer of reference gene in | CpActin-F | GTTATGGTTGGGATGGGACAGAAAG |
| CpActin-R | GGGCTTCAGTAAGGAAACAGGA | |
| Chimonanthus praecox | CpTublin-F | TAGTGACAAGACAGTAGGTGGAGGT |
| CpTublin-R | GTAGGTTCCAGTCCTCACTTCATC | |
| 拟南芥内参基因及内源基因实时荧光定量PCR qRT-PCR primer of reference and endogenous genes in Arabidopsis thaliana | AtActin-F | CTTCGTCTTCCACTTCAG |
| AtActin-R | ATCATACCAGTCTCAACAC | |
| AtLFY-F | AGACGGCTGCTTTTGGGATG | |
| AtLFY-R | TATTCCCCGCCGCATCAGTC | |
| AtFT -F | GATTGGTGGAGAAGACCTCAGGAAC | |
| AtFT -R | GCAGCCACTCTCCCTCTGACAAT | |
| AtAP1 -F | TAGGGCTCAACAGGAGCAGT | |
| AtAP1 -R | CAGCCAAGGTTGCAGTTGTA | |
| AtSOC1-F | AATTCGCCAGCTCCAATATG | |
| AtSOC1-R | CCTCGATTGAGCATGTTCCT |
表1 本研究所使用引物
Table 1 Primers used in this study
| 用途 Usage | 引物名称 Primer name | 序列(5′-3′) Sequence |
|---|---|---|
| CpAP2-L11基因克隆引物 CpAP2-L11 gene cloning primer | CpAP2-L11-F | AACAAATCGGACGGTTCAAACCACT |
| CpAP2-L11-R | GAAAATCAAAAACCGATCTCAGCCGG | |
| 表达载体构建引物 Expression vector construction primer | eCpAP2-L11-F | CGGGGTACCATGGGGAAACCATCACAGAAG |
| eCpAP2-L11-R | ACGCGTCGACTTACTGTCCATCCCAGTCGGC | |
| 实时荧光定量PCR qRT-PCR primer | qCpAP2-L11-F | TCCCGACCTTAACGTCTCGTCAGAG |
| qCpAP2-L11-R | GCAGCGGGGAGTTTTGTCGGATTCT | |
| 蜡梅内参基因实时荧光定量PCR qRT-PCR primer of reference gene in | CpActin-F | GTTATGGTTGGGATGGGACAGAAAG |
| CpActin-R | GGGCTTCAGTAAGGAAACAGGA | |
| Chimonanthus praecox | CpTublin-F | TAGTGACAAGACAGTAGGTGGAGGT |
| CpTublin-R | GTAGGTTCCAGTCCTCACTTCATC | |
| 拟南芥内参基因及内源基因实时荧光定量PCR qRT-PCR primer of reference and endogenous genes in Arabidopsis thaliana | AtActin-F | CTTCGTCTTCCACTTCAG |
| AtActin-R | ATCATACCAGTCTCAACAC | |
| AtLFY-F | AGACGGCTGCTTTTGGGATG | |
| AtLFY-R | TATTCCCCGCCGCATCAGTC | |
| AtFT -F | GATTGGTGGAGAAGACCTCAGGAAC | |
| AtFT -R | GCAGCCACTCTCCCTCTGACAAT | |
| AtAP1 -F | TAGGGCTCAACAGGAGCAGT | |
| AtAP1 -R | CAGCCAAGGTTGCAGTTGTA | |
| AtSOC1-F | AATTCGCCAGCTCCAATATG | |
| AtSOC1-R | CCTCGATTGAGCATGTTCCT |
| 名称Name | 框Scaffold | 位置Position | 氨基酸数Amino acids | 分子质量/kD Molecular weigh | 理论等电点pI |
|---|---|---|---|---|---|
| CpAP2-L1 | 8 | 633 532 ~ 638 961 | 593 | 65.68 | 6.27 |
| CpAP2-L2 | 12 | 6 666 416 ~ 6 670 043 | 501 | 55.82 | 6.29 |
| CpAP2-L3 | 18 | 2 386 489 ~ 2 389 495 | 516 | 57.15 | 6.06 |
| CpAP2-L4 | 18 | 4 068 635 ~ 4 073 270 | 650 | 71.47 | 7.14 |
| CpAP2-L5 | 22 | 197 714 ~ 200 051 | 389 | 43.87 | 7.12 |
| CpAP2-L6 | 26 | 6 547 145 ~ 6 551 504 | 447 | 49.63 | 5.88 |
| CpAP2-L7 | 32 | 2 029 444 ~ 2 034 037 | 643 | 71.22 | 6.22 |
| CpAP2-L8 | 70 | 2 623 638 ~ 2 627 212 | 479 | 54.38 | 6.17 |
| CpAP2-L9 | 72 | 5 070 756 ~ 5 074 288 | 460 | 50.50 | 7.63 |
| CpAP2-L10 | 86 | 4 575 244 ~ 4 577 540 | 394 | 44.47 | 6.62 |
| CpAP2-L11 | 92 | 926 637 ~ 932 737 | 345 | 38.67 | 6.11 |
| CpAP2-L12 | 120 | 4 203 257 ~ 4 209 225 | 650 | 71.21 | 6.32 |
| CpAP2-L13 | 122 | 2 412 620 ~ 2 417 460 | 721 | 78.19 | 6.06 |
| CpAP2-L14 | 140 | 668 231 ~ 673 284 | 403 | 45.23 | 8.38 |
| CpAP2-L15 | 153 | 849 837 ~ 855 009 | 686 | 75.31 | 6.02 |
| CpAP2-L16 | 156 | 1 808 018 ~ 1 817 462 | 472 | 52.60 | 6.61 |
| CpAP2-L17 | 243 | 654 864 ~ 688 642 | 356 | 40.11 | 7.67 |
| CpAP2-L18 | 463 | 3 517 149 ~ 3 524 013 | 496 | 54.80 | 6.03 |
| CpAP2-L19 | 742 | 586 727 ~ 591 191 | 396 | 43.87 | 5.51 |
| CpAP2-L20 | 752 | 2 116 052 ~ 2 120 180 | 477 | 51.37 | 8.07 |
表2 蜡梅AP2亚家族成员基本信息
Table 2 Basic information of the AP2 subfamily members in Chimonanthus praecox
| 名称Name | 框Scaffold | 位置Position | 氨基酸数Amino acids | 分子质量/kD Molecular weigh | 理论等电点pI |
|---|---|---|---|---|---|
| CpAP2-L1 | 8 | 633 532 ~ 638 961 | 593 | 65.68 | 6.27 |
| CpAP2-L2 | 12 | 6 666 416 ~ 6 670 043 | 501 | 55.82 | 6.29 |
| CpAP2-L3 | 18 | 2 386 489 ~ 2 389 495 | 516 | 57.15 | 6.06 |
| CpAP2-L4 | 18 | 4 068 635 ~ 4 073 270 | 650 | 71.47 | 7.14 |
| CpAP2-L5 | 22 | 197 714 ~ 200 051 | 389 | 43.87 | 7.12 |
| CpAP2-L6 | 26 | 6 547 145 ~ 6 551 504 | 447 | 49.63 | 5.88 |
| CpAP2-L7 | 32 | 2 029 444 ~ 2 034 037 | 643 | 71.22 | 6.22 |
| CpAP2-L8 | 70 | 2 623 638 ~ 2 627 212 | 479 | 54.38 | 6.17 |
| CpAP2-L9 | 72 | 5 070 756 ~ 5 074 288 | 460 | 50.50 | 7.63 |
| CpAP2-L10 | 86 | 4 575 244 ~ 4 577 540 | 394 | 44.47 | 6.62 |
| CpAP2-L11 | 92 | 926 637 ~ 932 737 | 345 | 38.67 | 6.11 |
| CpAP2-L12 | 120 | 4 203 257 ~ 4 209 225 | 650 | 71.21 | 6.32 |
| CpAP2-L13 | 122 | 2 412 620 ~ 2 417 460 | 721 | 78.19 | 6.06 |
| CpAP2-L14 | 140 | 668 231 ~ 673 284 | 403 | 45.23 | 8.38 |
| CpAP2-L15 | 153 | 849 837 ~ 855 009 | 686 | 75.31 | 6.02 |
| CpAP2-L16 | 156 | 1 808 018 ~ 1 817 462 | 472 | 52.60 | 6.61 |
| CpAP2-L17 | 243 | 654 864 ~ 688 642 | 356 | 40.11 | 7.67 |
| CpAP2-L18 | 463 | 3 517 149 ~ 3 524 013 | 496 | 54.80 | 6.03 |
| CpAP2-L19 | 742 | 586 727 ~ 591 191 | 396 | 43.87 | 5.51 |
| CpAP2-L20 | 752 | 2 116 052 ~ 2 120 180 | 477 | 51.37 | 8.07 |
| 基因1 Gene 1 | 基因2 Gene 2 | 非同义 替换率Ka | 同义 替换率Ks | Ka/Ks | 选择类型 Selection type | 复制类型 Duplication type |
|---|---|---|---|---|---|---|
| CpAP2-L1 | CpAP2-L11 | 0.59 | NaN | NaN | 纯化选择Purify selection | 全基因组复制或片段复制WGD or SD |
| CpAP2-L3 | CpAP2-L4 | 0.78 | NaN | NaN | 纯化选择Purify selection | 全基因组复制或片段复制WGD or SD |
| CpAP2-L3 | CpAP2-L8 | 0.20 | 0.66 | 0.30 | 纯化选择Purify selection | 全基因组复制或片段复制WGD or SD |
| CpAP2-L3 | CpAP2-L11 | 0.66 | NaN | NaN | 纯化选择Purify selection | 全基因组复制或片段复制WGD or SD |
| CpAP2-L4 | CpAP2-L7 | 0.19 | 0.82 | 0.24 | 纯化选择Purify selection | 全基因组复制或片段复制WGD or SD |
| CpAP2-L4 | CpAP2-L1 | 0.18 | 1.02 | 0.17 | 纯化选择Purify selection | 全基因组复制或片段复制WGD or SD |
| CpAP2-L6 | CpAP2-L11 | 0.67 | NaN | NaN | 纯化选择Purify selection | 全基因组复制或片段复制WGD or SD |
| CpAP2-L8 | CpAP2-L11 | 0.66 | NaN | NaN | 纯化选择Purify selection | 全基因组复制或片段复制WGD or SD |
| CpAP2-L12 | CpAP2-L3 | 0.72 | NaN | NaN | 纯化选择Purify selection | 全基因组复制或片段复制WGD or SD |
| CpAP2-L12 | CpAP2-L4 | 0.51 | 2.37 | 0.22 | 纯化选择Purify selection | 全基因组复制或片段复制WGD or SD |
| CpAP2-L12 | CpAP2-L8 | 0.66 | NaN | NaN | 纯化选择Purify selection | 全基因组复制或片段复制WGD or SD |
| CpAP2-L12 | CpAP2-L11 | 0.51 | NaN | NaN | 纯化选择Purify selection | 全基因组复制或片段复制WGD or SD |
| CpAP2-L14 | CpAP2-L10 | 0.16 | 0.73 | 0.22 | 纯化选择Purify selection | 全基因组复制或片段复制WGD or SD |
| CpAP2-L14 | CpAP2-L11 | 0.49 | NaN | NaN | 纯化选择Purify selection | 全基因组复制或片段复制WGD or SD |
| CpAP2-L15 | CpAP2-L11 | 0.55 | 2.48 | 0.22 | 纯化选择Purify selection | 全基因组复制或片段复制WGD or SD |
| CpAP2-L16 | CpAP2-L10 | 0.58 | NaN | NaN | 纯化选择Purify selection | 全基因组复制或片段复制WGD or SD |
表3 蜡梅AP2亚家族成员进化选择类型及复制类型
Table 3 Evolutionary selection types and duplication types of the AP2 subfamily members in Chimonanthus praecox
| 基因1 Gene 1 | 基因2 Gene 2 | 非同义 替换率Ka | 同义 替换率Ks | Ka/Ks | 选择类型 Selection type | 复制类型 Duplication type |
|---|---|---|---|---|---|---|
| CpAP2-L1 | CpAP2-L11 | 0.59 | NaN | NaN | 纯化选择Purify selection | 全基因组复制或片段复制WGD or SD |
| CpAP2-L3 | CpAP2-L4 | 0.78 | NaN | NaN | 纯化选择Purify selection | 全基因组复制或片段复制WGD or SD |
| CpAP2-L3 | CpAP2-L8 | 0.20 | 0.66 | 0.30 | 纯化选择Purify selection | 全基因组复制或片段复制WGD or SD |
| CpAP2-L3 | CpAP2-L11 | 0.66 | NaN | NaN | 纯化选择Purify selection | 全基因组复制或片段复制WGD or SD |
| CpAP2-L4 | CpAP2-L7 | 0.19 | 0.82 | 0.24 | 纯化选择Purify selection | 全基因组复制或片段复制WGD or SD |
| CpAP2-L4 | CpAP2-L1 | 0.18 | 1.02 | 0.17 | 纯化选择Purify selection | 全基因组复制或片段复制WGD or SD |
| CpAP2-L6 | CpAP2-L11 | 0.67 | NaN | NaN | 纯化选择Purify selection | 全基因组复制或片段复制WGD or SD |
| CpAP2-L8 | CpAP2-L11 | 0.66 | NaN | NaN | 纯化选择Purify selection | 全基因组复制或片段复制WGD or SD |
| CpAP2-L12 | CpAP2-L3 | 0.72 | NaN | NaN | 纯化选择Purify selection | 全基因组复制或片段复制WGD or SD |
| CpAP2-L12 | CpAP2-L4 | 0.51 | 2.37 | 0.22 | 纯化选择Purify selection | 全基因组复制或片段复制WGD or SD |
| CpAP2-L12 | CpAP2-L8 | 0.66 | NaN | NaN | 纯化选择Purify selection | 全基因组复制或片段复制WGD or SD |
| CpAP2-L12 | CpAP2-L11 | 0.51 | NaN | NaN | 纯化选择Purify selection | 全基因组复制或片段复制WGD or SD |
| CpAP2-L14 | CpAP2-L10 | 0.16 | 0.73 | 0.22 | 纯化选择Purify selection | 全基因组复制或片段复制WGD or SD |
| CpAP2-L14 | CpAP2-L11 | 0.49 | NaN | NaN | 纯化选择Purify selection | 全基因组复制或片段复制WGD or SD |
| CpAP2-L15 | CpAP2-L11 | 0.55 | 2.48 | 0.22 | 纯化选择Purify selection | 全基因组复制或片段复制WGD or SD |
| CpAP2-L16 | CpAP2-L10 | 0.58 | NaN | NaN | 纯化选择Purify selection | 全基因组复制或片段复制WGD or SD |
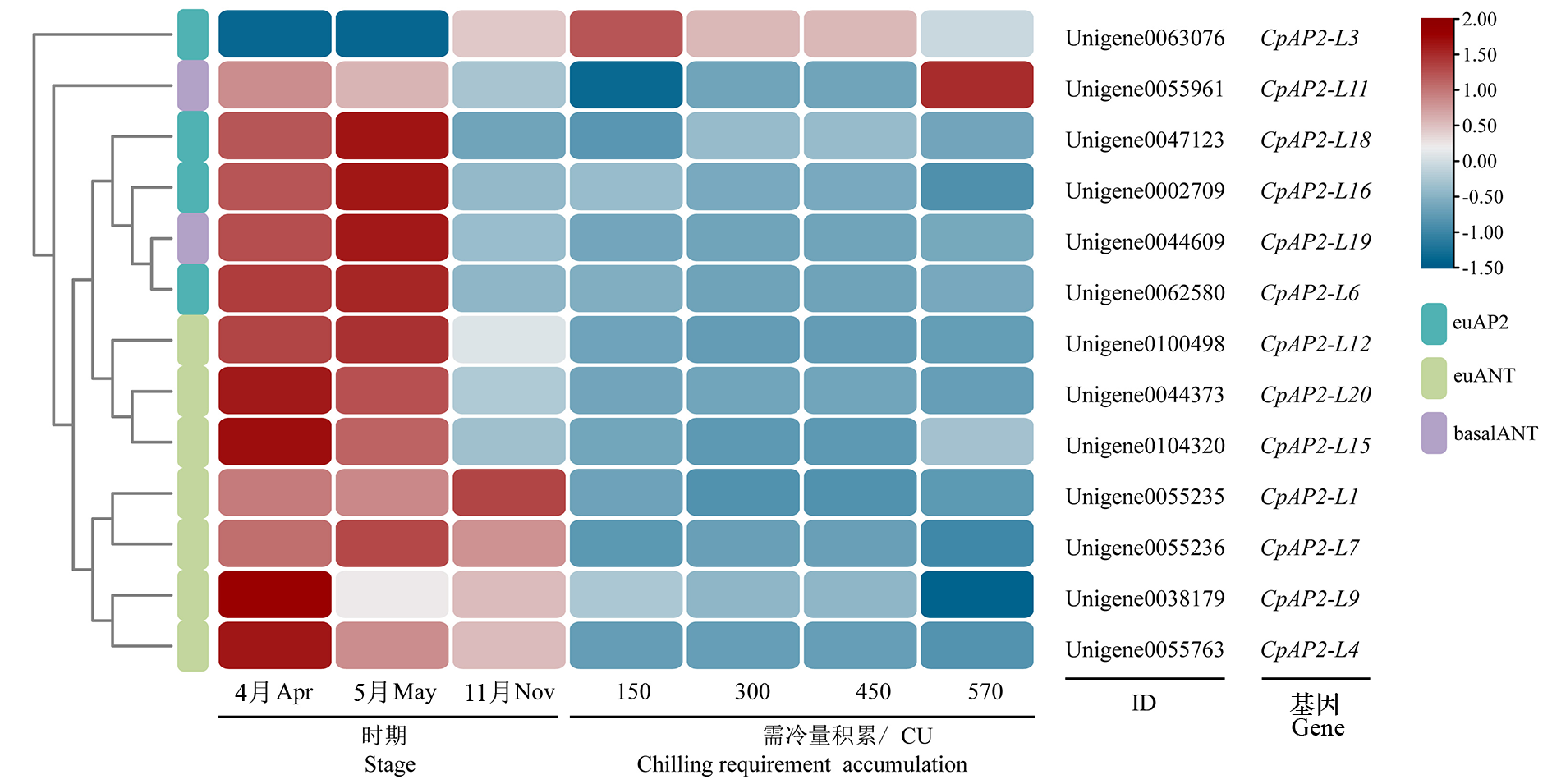
图4 花发育不同时期及不同需冷量积累下蜡梅AP2亚家族成员表达热图 570为始花期花芽,需冷量积累570 CU。
Fig. 4 Heatmap of the expression of members of the AP2 subfamily in Chimonanthus praecox at different stages of flower development and different chilling requirements 570 indicates the flower bud at beginning of initiating blooming stage.
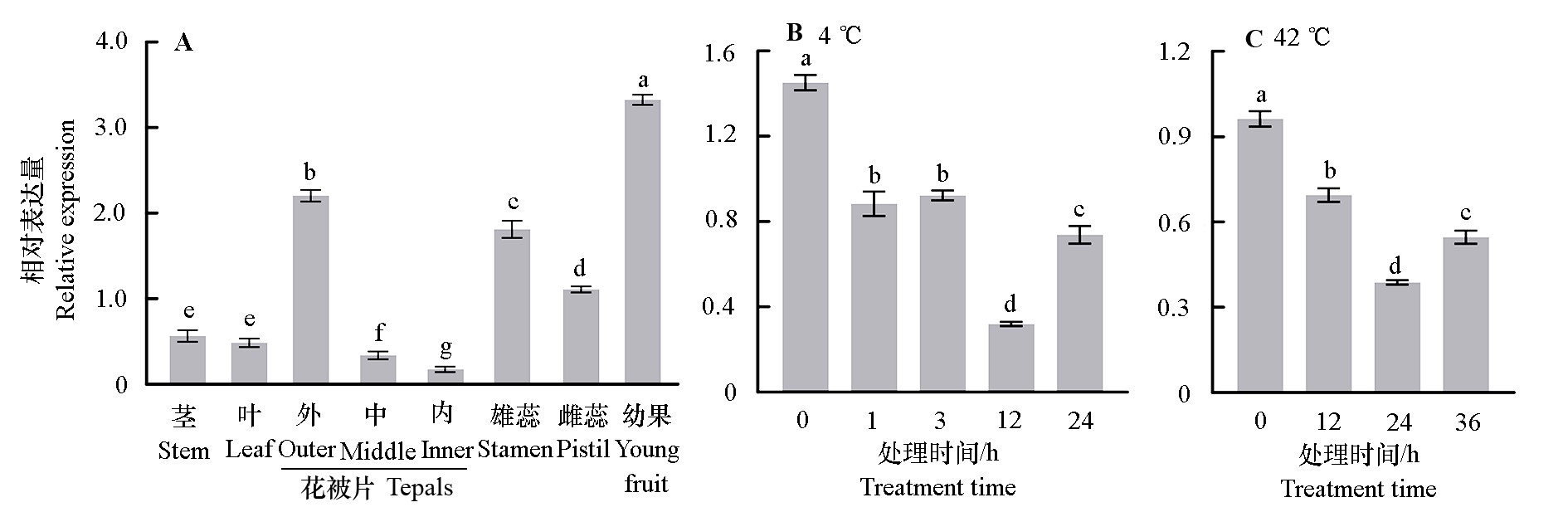
图5 蜡梅不同组织、器官(A)及温度条件下(B,C)CpAP2-L11相对表达量 不同小写字母表示差异显著(P < 0.05)。下同。
Fig. 5 Relative expression of CpAP2-L11 in different tissues,organs(A),and temperature conditions(B,C)in Chimonanthus praecox Different lowercase letters indicate significant differences(P < 0.05). The same below.
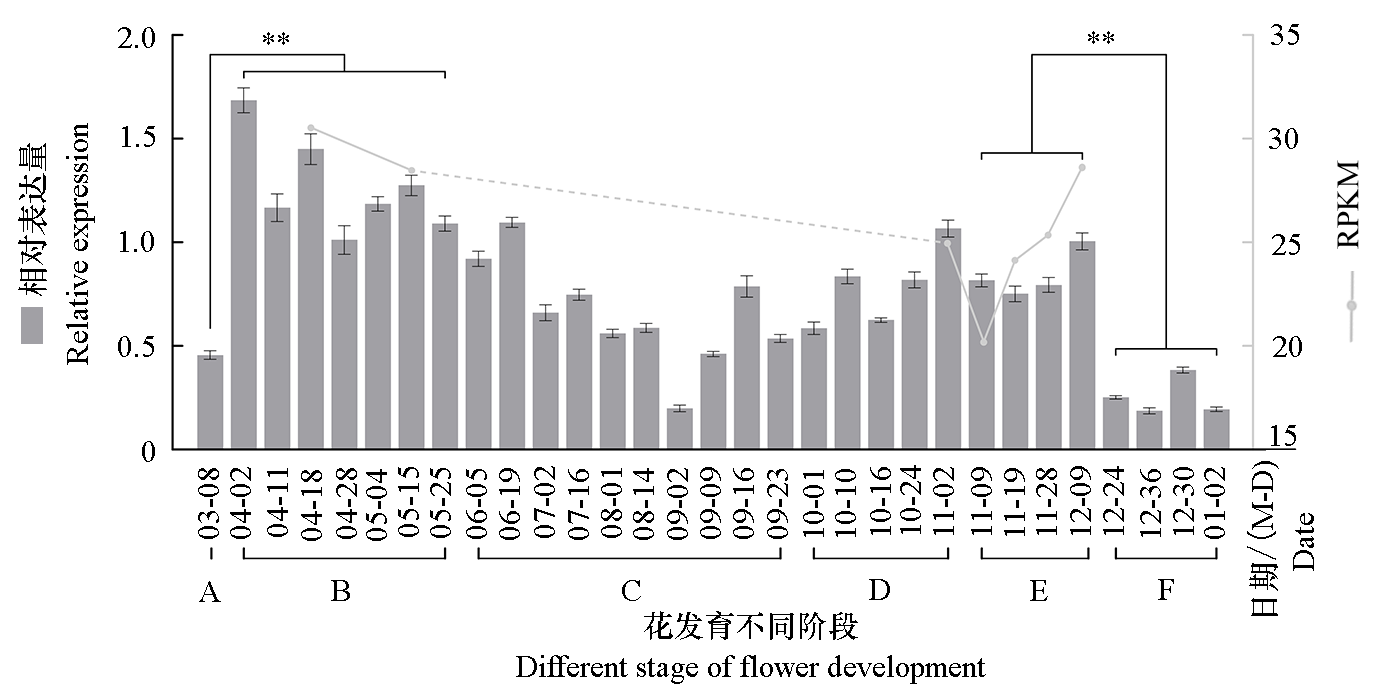
图6 蜡梅花发育不同时期(柱状图)及转录组数据库中(折线图)CpAP2-L11相对表达量 A:枝芽阶段;B:花原基形成、雌/雄蕊分化阶段;C:花芽休眠阶段;D:子房发育阶段;E:需冷量积累阶段;F:花蕾开放。** 表示具有显著差异(P < 0.05)。
Fig. 6 Relative expression of CpAP2-L11 in different stages of the flower development in Chimonanthus praecox(histogram)and transcriptome database(line graph) A:Branch bud;B:Flower primordium formation and stamen/pistil differentiation;C:Flower bud primordium dormancy;D:Ovary development;E:Chilling requirements accumulation;F:Flower buds opening. ** indicate significant differences(P < 0.05).

图7 拟南芥CpAP2-L11过表达T2代株系(OE)的CpAP2-L11相对表达量(A)和表型(B)
Fig. 7 Relative expression of CpAP2-L11(A)and phenotype(B)in over expression T2 generation lines of Arabidopsis
| 株系Line | 莲座叶数 Number of rosette leaves | 时间/d Time | 株高/cm Plant height | ||
|---|---|---|---|---|---|
| 抽薹1 cm Bolting 1 cm | 第1朵花开放 The first flower oping | 第1个果荚形成 The first pod formation | |||
| 野生型Wild type | 10.3 ± 0.9 a | 27.4 ± 0.9 a | 30.6 ± 1.2 a | 33.8 ± 1.1 a | 25.04 ± 0.81 a |
| OE10-12 | 8.3 ± 0.9 b | 25.5 ± 1.2 b | 28.4 ± 1.4 b | 30.9 ± 1.6 b | 18.88 ± 0.88 b |
| OE9-5 | 7.9 ± 0.7 bc | 25.0 ± 1.0 b | 27.8 ± 1.0 b | 30.3 ± 0.9 b | 18.48 ± 0.47 b |
| OE7-6 | 7.4 ± 0.6 c | 22.6 ± 1.1 c | 25.6 ± 1.2 c | 28.3 ± 1.4 c | 17.06 ± 0.55 c |
表4 拟南芥CpAP2-L11过表达株系(OE)的表型
Table 4 Phenotype of over expression lines in 35S::CpAP2-L11 Arabidopsis
| 株系Line | 莲座叶数 Number of rosette leaves | 时间/d Time | 株高/cm Plant height | ||
|---|---|---|---|---|---|
| 抽薹1 cm Bolting 1 cm | 第1朵花开放 The first flower oping | 第1个果荚形成 The first pod formation | |||
| 野生型Wild type | 10.3 ± 0.9 a | 27.4 ± 0.9 a | 30.6 ± 1.2 a | 33.8 ± 1.1 a | 25.04 ± 0.81 a |
| OE10-12 | 8.3 ± 0.9 b | 25.5 ± 1.2 b | 28.4 ± 1.4 b | 30.9 ± 1.6 b | 18.88 ± 0.88 b |
| OE9-5 | 7.9 ± 0.7 bc | 25.0 ± 1.0 b | 27.8 ± 1.0 b | 30.3 ± 0.9 b | 18.48 ± 0.47 b |
| OE7-6 | 7.4 ± 0.6 c | 22.6 ± 1.1 c | 25.6 ± 1.2 c | 28.3 ± 1.4 c | 17.06 ± 0.55 c |
| [1] | Ahmed S, Rashid M A R, Zafar S A, Azhar M T, Waqas M, Uzair M, Rana I A, Azeem F, Chung G, Ali Z, Atif R M. 2021. Genome-wide investigation and expression analysis of APETALA-2 transcription factor subfamily reveals its evolution,expansion and regulatory role in abiotic stress responses in Indica rice(Oryza sativa L. ssp. indica). Genomics, 113 (1 Pt 2):10291043. |
| [2] |
Aukerman M J, Sakai H. 2003. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-Like target genes. The Plant Cell, 15 (11):2730-2741.
doi: 10.1105/tpc.016238 URL |
| [3] | Bennetzen J L. 2005. Transposable elements,gene creation and genome rearrangement in flowering plants. Current Opinion in Genetics & Development, 15 (6):621-627. |
| [4] |
Boutilier K, Offringa R, Sharma V K, Kieft H, Ouellet T, Zhang L, Hattori J, Liu C M, van Lammeren A A M, Miki B L A, Custers J B M, van Lookeren Campagne M M. 2002. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. The Plant Cell, 14 (8):1737-1749.
doi: 10.1105/tpc.001941 URL |
| [5] |
Cannon S B, Mitra A, Baumgarten A, Young N D, May G. 2004. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biology, 4 (1):10.
doi: 10.1186/1471-2229-4-10 URL |
| [6] |
Chen C J, Chen H, Zhang Y, Thomas H R, Frank M H, He Y, Xia R. 2020. Tbtools:an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 13 (8):1194-1202.
doi: 10.1016/j.molp.2020.06.009 URL |
| [7] |
Clough S J, Bent A F. 1998. Floral dip:a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant Journal, 16 (6):735-743.
doi: 10.1046/j.1365-313x.1998.00343.x pmid: 10069079 |
| [8] | Cui bo, Hao Ping’an, Liang Fang, Zhang Yan, Wang Ximeng, Li Junlin, Jiang Suhua, Xu Shenping. 2020. Cloning and expression analysis of AP2/ERF family gene from Phalaenopsis under low temperature. Acta Horticulturae Sinica, 47 (1):85-97. (in Chinese) |
|
崔波, 郝平安, 梁芳, 张燕, 王喜蒙, 李俊霖, 蒋素华, 许申平. 2020. 蝴蝶兰AP2/ERF家族基因的克隆及在低温下表达特性分析. 园艺学报, 47 (1):85-97.
doi: 10.16420/j.issn.0513-353x.2019-0091 |
|
| [9] |
Dong C, Xi Y, Chen X, Cheng Z M. 2021. Genome-wide identification of AP2/EREBP in Fragaria vesca and expression pattern analysis of the FvDREB subfamily under drought stress. BMC Plant Biology, 21 (1):295.
doi: 10.1186/s12870-021-03095-2 |
| [10] |
Faraji S, Filiz E, Kazemitabar S K, Vannozzi A, Palumbo F, Barcaccia G, Heidari P. 2020. The AP2/ERF gene family in Triticum durum:genome-wide identification and expression analysis under drought and salinity stresses. Genes, 11 (12):1464.
doi: 10.3390/genes11121464 URL |
| [11] |
Feng K, Hou X L, Xing G M, Liu J X, Duan A Q, Xu Z S, Li M Y, Zhuang J, Xiong A S. 2020. Advances in AP2/ERF super-family transcription factors in plant. Critical Reviews in Biotechnology, 40 (6):750-776.
doi: 10.1080/07388551.2020.1768509 pmid: 32522044 |
| [12] | Green M R, Sambrook J. 2001. Molecular cloning:a laboratory manual. Analytical Biochemistry, 186 (1):182-183. |
| [13] |
Gutterson N, Reuber T L. 2004. Regulation of disease resistance pathways by AP2/ERF transcription factors. Current Opinion in Plant Biology, 7 (4):465-471.
doi: 10.1016/j.pbi.2004.04.007 pmid: 15231271 |
| [14] |
Jiang F, Guo M, Yang F, Duncan K, Jackson D, Rafalski A, Wang S, Li B. 2012. Mutations in an AP2 transcription factor-like gene affect internode length and leaf shape in maize. PLoS ONE, 7 (5):e37040.
doi: 10.1371/journal.pone.0037040 URL |
| [15] |
Jin J H, Wang M, Zhang H X, Khan A, Wei A M, Luo D X, Gong Z H, Tar’an B. 2018. Genome-wide identification of the AP2/ERF transcription factor family in pepper(Capsicum annuum L.). Genome, 61 (9):663-674.
doi: 10.1139/gen-2018-0036 URL |
| [16] |
Jose L R, Elliot M M. 1998. The AP2/EREBP family of plant transcription factors. Biological Chemistry, 379 (6):633-654.
doi: 10.1515/bchm.1998.379.6.633 pmid: 9687012 |
| [17] | Jung J H, Lee S, Yun J, Lee M, Park C M. 2014. The miR172 target TOE3 represses AGAMOUS expression during Arabidopsis floral patterning. Plant Science, 215:29-38. |
| [18] |
Kim S, Soltis P S, Wall K, Soltis D E. 2006. Phylogeny and domain evolution in the APETALA2-like gene family. Molecular Biology and Evolution, 23 (1):107-120.
pmid: 16151182 |
| [19] | Kondrashov F A, Rogozin I B, Wolf Y I, Koonin E V. 2002. Selection in the evolution of gene duplications. Genome Biology, 3 (2):research0008.0001. |
| [20] |
Krizek B A. 2011. Aintegumenta and Aintegumenta-Like6 regulate auxin-mediated flower development in Arabidopsis. BMC Research Notes, 4 (1):176.
doi: 10.1186/1756-0500-4-176 |
| [21] |
Krizek B A, Eaddy M. 2012. AINTEGUMENTA-LIKE6 regulates cellular differentiation in flowers. Plant Molecular Biology, 78 (3):199-209.
doi: 10.1007/s11103-011-9844-3 pmid: 22076630 |
| [22] |
Lecharny A, Boudet N, Gy I, Aubourg S, Kreis M. 2003. Introns in,introns out in plant gene families:a genomic approach of the dynamics of gene structure. J Struct Funct Genomics, 3 (1-4):111-116.
doi: 10.1023/A:1022614001371 URL |
| [23] |
Lei M, Li Z Y, Wang J B, Fu Y L, Xu L. 2019. Ectopic expression of the Aechmea fasciata APETALA2 gene AfAP2-2 reduces seed size and delays flowering in Arabidopsis. Plant Physiology and Biochemistry, 139:642-650.
doi: 10.1016/j.plaphy.2019.03.034 URL |
| [24] |
Li A, Yu X, Cao B B, Peng L X, Gao Y, Feng T, Li H, Ren Z Y. 2017. LkAP2L2,an AP2/ERF transcription factor gene of Larix kaempferi,with pleiotropic roles in plant branch and seed development. Russian Journal of Genetics, 53 (12):1335-1342.
doi: 10.1134/S1022795417120079 URL |
| [25] |
Li J, Chen F, Li Y, Li P, Wang Y, Mi G, Yuan L. 2019. ZmRAP2.7,an AP 2 transcription factor, is involved in Maize brace roots development. Frontiers in Plant Science, 10 (1):820.
doi: 10.3389/fpls.2019.00820 URL |
| [26] |
Li Ying, Meng Xianwei, Ma Zhihang, Liu Mengjun, Zhao Jin. 2022. Identification and expression analysis of microRNA families associated with phase transition in Chinese jujube. Acta Horticulturae Sinica, 49 (1):23-40. (in Chinese)
doi: 10.16420/j.issn.0513-353x.2020-0969 |
|
李莹, 孟宪巍, 马志航, 刘孟军, 赵锦. 2022. 枣树阶段转变相关microRNA家族的鉴定及其表达分析. 园艺学报, 49 (1):23-40.
doi: 10.16420/j.issn.0513-353x.2020-0969 |
|
| [27] |
Li Z N, Liu N, Zhang W, Wu C, Jiang Y, Ma J, Li M, Sui S. 2020. Integrated transcriptome and proteome analysis provides insight into chilling-induced dormancy breaking in Chimonanthus praecox. Horticulture Research, 7 (1):198.
doi: 10.1038/s41438-020-00421-x |
| [28] |
Licausi F, Giorgi F M, Zenoni S, Osti F, Pezzotti M, Perata P. 2010. Genomic and transcriptomic analysis of the AP2/ERF superfamily in Vitis vinifera. BMC Genomics, 11 (1):719.
doi: 10.1186/1471-2164-11-719 URL |
| [29] |
Licausi F, Ohme-Takagi M, Perata P. 2013. APETALA2/ethylene responsive factor(AP2/ERF)transcription factors:mediators of stress responses and developmental programs. New Phytologist, 199 (3):639-649.
doi: 10.1111/nph.12291 URL |
| [30] |
Liu M, Sun W, Ma Z, Zheng T, Huang L, Wu Q, Zhao G, Tang Z, Bu T, Li C, Chen H. 2019. Genome-wide investigation of the AP2/ERF gene family in tartary buckwheat(Fagopyum tataricum). BMC Plant Biology, 19:84.
doi: 10.1186/s12870-019-1681-6 |
| [31] |
Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆CT method. Methods, 25 (4):402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [32] |
Ma Xinrui, Li Liang, Liu Jinhang, Yang Mengjie, Chen Jie, Liang Qin, Wu Shaohua, Li Yongyu. 2018. Identification and differentially expressed analysis of microRNA associated with dormancy of pear flower buds. Acta Horticulturae Sinica, 45 (11):2089-2105. (in Chinese)
doi: 10.16420/j.issn.0513-353x.2018-0012 |
|
马鑫瑞, 李亮, 刘瑾航, 杨梦洁, 陈洁, 梁沁, 吴少华, 李永裕. 2018. 梨花芽休眠相关miRNA的鉴定和差异表达分析. 园艺学报, 45 (11):2089-2105.
doi: 10.16420/j.issn.0513-353x.2018-0012 |
|
| [33] |
Maes T, van de Steene N, Zethof J, Karimi M, D'Hauw M, Mares G, van Montagu M, Gerats T. 2001. Petunia AP2-like genes and their role in flower and seed development. The Plant Cell, 13 (2):229-244.
doi: 10.1105/tpc.13.2.229 URL |
| [34] |
Najafi S, Sorkheh K, Nasernakhaei F. 2018. Characterization of the APETALA2/ethylene-responsive factor(AP2/ERF)transcription factor family in sunflower. Scientific Reports, 8:11576.
doi: 10.1038/s41598-018-29526-z |
| [35] |
Nakano T, Suzuki K, Fujimura T, Shinshi H. 2006. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiology, 140 (2):411-432.
doi: 10.1104/pp.105.073783 URL |
| [36] |
Nilsson L, Carlsbecker A, Sundås-Larsson A, Vahala T. 2007. APETALA2 like genes from Picea abies show functional similarities to their Arabidopsis homologues. Planta, 225 (3):589-602.
pmid: 16953432 |
| [37] |
O'Maoileidigh D S, van Driel A D, Singh A, Sang Q,le Bec N, Vincent C, de Olalla E B G, Vayssieres A, Romera Branchat M, Severing E, Martinez Gallegos R, Coupland G. 2021. Systematic analyses of the MIR172 family members of Arabidopsis define their distinct roles in regulation of APETALA2 during floral transition. PLoS Biology, 19 (2):e3001043.
doi: 10.1371/journal.pbio.3001043 URL |
| [38] |
Park W, Li J, Song R, Messing J, Chen X. 2002. CARPEL FACTORY,a dicer homolog,and HEN1,a novel protein,zct in microRNA metabolism in Arabidopsis thaliana. Current Biology, 12 (17):1484-1495.
doi: 10.1016/S0960-9822(02)01017-5 URL |
| [39] | Riaz M W, Lu J, Shah L, Yang L, Chen C, Mei X D, Xue L, Manzoor M A, Abdullah M, Rehman S, Si H, Ma C. 2021. Expansion and molecular characterization of AP2/ERF gene family in wheat(Triticum aestivum L.). Frontiers in Genetics, 12:Article 632155. |
| [40] |
Riechmann J L, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe O J, Samaha R R, Creelman R, Pilgrim M, Broun P, Zhang J Z, Ghandehari D, Sherman B K, Yu G. 2000. Arabidopsis transcription factors:genome-wide comparative analysis among eukaryotes. Science, 290 (5499):2105-2115.
doi: 10.1126/science.290.5499.2105 pmid: 11118137 |
| [41] |
Roy S W, Gilbert W. 2005. Rates of intron loss and gain:implications for early eukaryotic evolution. Proc Natl Acad Sci U S A, 102 (16):5773.
doi: 10.1073/pnas.0500383102 URL |
| [42] |
Shang J, Tian J, Cheng H, Yan Q, Li L, Jamal A, Xu Z, Xiang L, Saski C A, Jin S, Zhao K, Liu X, Chen L. 2020. The chromosome-level wintersweet(Chimonanthus praecox)genome provides insights into floral scent biosynthesis and flowering in winter. Genome Biology, 21 (1):200.
doi: 10.1186/s13059-020-02088-y |
| [43] |
Wang R, Cheng Y, Ke X, Zhang X, Zhang H, Huang J. 2020. Comparative analysis of salt responsive gene regulatory networks in rice and Arabidopsis. Computational Biology and Chemistry, 85 (1):107188.
doi: 10.1016/j.compbiolchem.2019.107188 URL |
| [44] |
Wang Y, Tang H, Debarry J D, Tan X, Li J, Wang X, Lee T H, Jin H, Marler B, Guo H, Kissinger J C, Paterson A H. 2012. MCScanX:a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Research, 40 (7):e49.
doi: 10.1093/nar/gkr1293 URL |
| [45] |
Wollmann H, Mica E, Todesco M, Long J A, Weigel D. 2010. On reconciling the interactions between APETALA2,miR172 and AGAMOUS with the ABC model of flower development. Development, 137 (21):3633-3642.
doi: 10.1242/dev.036673 pmid: 20876650 |
| [46] |
Xie Y, Yu X, Jiang S, Xiao K, Wang Y, Li L, Wang F, He W, Cai Q, Xie H, Zhang J. 2020. OsGL6,a conserved AP 2 domain protein,promotes leaf trichome initiation in rice. Biochemical and Biophysical Research Communications, 522 (2):448-455.
doi: 10.1016/j.bbrc.2019.11.125 URL |
| [47] |
Xie Z, Nolan T M, Jiang H, Yin Y. 2019. AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Frontiers in Plant Science, 10:228.
doi: 10.3389/fpls.2019.00228 URL |
| [48] |
Zhang B, Wang L, Zeng L, Zhang C, Ma H. 2015. Arabidopsis TOE proteins convey a photoperiodic signal to antagonize CONSTANS and regulate flowering time. Genes & Development, 29 (9):975-987.
doi: 10.1101/gad.251520.114 URL |
| [49] |
Xu Hongxia, Zhou Huifen, Li Xiaoying, Jiang Luhua, Chen Junwei. 2021. Comparative transcriptome analysis of different developmental stages of flowers and fruits in loquat under low temperature stress. Acta Horticulturae Sinica, 48 (9):1680-1694. (in Chinese)
doi: 10.16420/j.issn.0513-353x.2021- 0219 |
|
徐红霞, 周慧芬, 李晓颖, 姜路花, 陈俊伟. 2021. 低温胁迫下枇杷不同发育阶段的花果转录组比较分析. 园艺学报, 48 (9):1680-1694.
doi: 10.16420/j.issn.0513-353x.2021- 0219 |
|
| [50] |
Zhang J, Shi S Z, Jiang Y, Zhong F, Liu G, Yu C, Lian B, Chen Y. 2021. Genome-wide investigation of the AP2/ERF superfamily and their expression under salt stress in Chinese willow(Salix matsudana). Peerj, 9 (1):e11076.
doi: 10.7717/peerj.11076 URL |
| [51] |
Zhang S T, Zhu C, Lyu Y M, Chen Y, Zhang Z H, Lai Z X, Lin Y L. 2020. Genome-wide identification,molecular evolution,and expression analysis provide new insights into the APETALA2/ethylene responsive factor(AP2/ERF)superfamily in Dimocarpus longan Lour. BMC Genomics, 21 (1):62.
doi: 10.1186/s12864-020-6469-4 |
| [52] |
Zhou L, Yarra R. 2021. Genome-wide identification and characterization of AP2/ERF transcription factor family genes in oil palm under abiotic stress conditions. International Journal of Molecular Sciences, 22 (6):2821.
doi: 10.3390/ijms22062821 URL |
| [53] | Zhu P, Chen Y, Zhang J, Wu F, Wang X, Pan T, Wei Q, Hao Y, Chen X, Jiang C, Ji K. 2021. Identification,classification,and characterization of AP2/ERF superfamily genes in Masson pine(Pinus massoniana Lamb.). Scientific Report, 11 (1):5441. |
| [54] |
Zhuang J, Xiong A S, Peng R H, Gao F, Zhu B, Zhang J, Fu X Y, Jin X F, Chen J M, Zhang Z, Qiao Y S, Yao Q H. 2010. Analysis of Brassica rapa ESTs:gene discovery and expression patterns of AP2/ERF family genes. Molecular Biology Reports, 37 (5):2485-2492.
doi: 10.1007/s11033-009-9763-4 pmid: 19701799 |
| [1] | 刘语诺, 曹亚, 王帅, 杜美霞, 郑林, 陈善春, 邹修平. 柑橘CsMYB41和CsMYB63响应溃疡病菌侵染的表达[J]. 园艺学报, 2023, 50(3): 495-507. |
| [2] | 叶子茂, 申晚霞, 刘梦雨, 王彤, 张晓楠, 余歆, 刘小丰, 赵晓春. R2R3-MYB转录因子CitMYB21对柑橘类黄酮生物合成的影响[J]. 园艺学报, 2023, 50(2): 250-264. |
| [3] | 宋艳红, 陈亚铎, 张晓玉, 宋盼, 刘丽锋, 李刚, 赵霞, 周厚成. 森林草莓FvbHLH130转录因子调控植株提前开花[J]. 园艺学报, 2023, 50(2): 295-306. |
| [4] | 韩睿, 钟雄辉, 陈登辉, 崔建, 乐祥庆, 颉建明, 康俊根. 黑腐病菌效应因子XopR的甘蓝靶标基因BobHLH34的克隆及功能分析[J]. 园艺学报, 2023, 50(2): 319-330. |
| [5] | 任菲, 卢苗苗, 刘吉祥, 陈信立, 刘道凤, 眭顺照, 马婧. 蜡梅胚胎晚期丰富蛋白基因CpLEA的表达及抗性分析[J]. 园艺学报, 2023, 50(2): 359-370. |
| [6] | 蔺海娇, 梁雨晨, 李玲, 马军, 张璐, 兰振颖, 苑泽宁. 薰衣草CBF途径相关耐寒基因挖掘与调控网络分析[J]. 园艺学报, 2023, 50(1): 131-144. |
| [7] | 贾鑫, 曾臻, 陈月, 冯慧, 吕英民, 赵世伟. 月季‘月月粉’RcDREB2A的克隆与表达分析[J]. 园艺学报, 2022, 49(9): 1945-1956. |
| [8] | 宋兴荣, 袁蒲英, 何相达. 蜡梅新品种‘变早素’[J]. 园艺学报, 2022, 49(9): 2059-2060. |
| [9] | 许海峰, 王中堂, 陈新, 刘志国, 王利虎, 刘平, 刘孟军, 张琼. 冬枣果皮着色相关类黄酮靶向代谢组学及潜在MYB转录因子分析[J]. 园艺学报, 2022, 49(8): 1761-1771. |
| [10] | 郑林, 王帅, 刘语诺, 杜美霞, 彭爱红, 何永睿, 陈善春, 邹修平. 柑橘响应黄龙病菌侵染的NAC基因的克隆及表达分析[J]. 园艺学报, 2022, 49(7): 1441-1457. |
| [11] | 钱婕妤, 蒋玲莉, 郑钢, 陈佳红, 赖吴浩, 许梦晗, 付建新, 张超. 百日草花青素苷合成相关MYB转录因子筛选及ZeMYB9功能研究[J]. 园艺学报, 2022, 49(7): 1505-1518. |
| [12] | 陈道宗, 刘镒, 沈文杰, 朱博, 谭晨. 白菜、甘蓝和甘蓝型油菜PAP1/2同源基因的鉴定及分析[J]. 园艺学报, 2022, 49(6): 1301-1312. |
| [13] | 王妍, 孙政, 冯珊, 袁心怡, 仲林林, 曾云流, 傅小鹏, 程运江, 包满珠, 张帆. 香石竹DcERF-1转录因子对切花衰老的负调控作用[J]. 园艺学报, 2022, 49(6): 1313-1326. |
| [14] | 沈植国, 张琳, 袁德义, 程建明. 蜡梅花色及其红花新资源研究进展[J]. 园艺学报, 2022, 49(4): 924-934. |
| [15] | 黎春红, 汪开拓, 雷长毅, 许凤, 季娜娜, 蒋永波. 桃TGA家族鉴定及BABA诱导的抗病表达分析[J]. 园艺学报, 2022, 49(2): 265-280. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司