
园艺学报 ›› 2023, Vol. 50 ›› Issue (1): 65-78.doi: 10.16420/j.issn.0513-353x.2021-0985
崔建1,2, 钟雄辉2, 刘泽慈2, 陈登辉2, 李海龙2, 韩睿2, 乐祥庆2, 康俊根2,*( ), 王超1,*(
), 王超1,*( )
)
收稿日期:2022-08-17
修回日期:2022-11-17
出版日期:2023-01-25
发布日期:2023-01-18
通讯作者:
*(E-mail:基金资助:
CUI Jian1,2, ZHONG Xionghui2, LIU Zeci2, CHEN Denghui2, LI Hailong2, HAN Rui2, YUE Xiangqing2, KANG Jungen2,*( ), WANG Chao1,*(
), WANG Chao1,*( )
)
Received:2022-08-17
Revised:2022-11-17
Online:2023-01-25
Published:2023-01-18
Contact:
*(E-mail:摘要:
以结球甘蓝(Brassica oleracea L. var. capitata)育种骨干亲本R2-P2(圆球形,抽薹晚,硫苷含量低,高感黑腐病、枯萎病和霜霉病)为轮回亲本,以野生甘蓝自交系R4-P1(不结球,抽薹早,蜡质厚,硫苷含量高,对黑腐病、枯萎病和霜霉病等多种病害具有抗性)为供体亲本,通过连续回交、自交和分子标记辅助选择,构建出一套由163个具有R2-P2遗传背景且具有R4-P1供体染色体片段单株组成的染色体片段替换系(Chromosome segment substitution line)。该替换系群体具有102个染色体替换片段,均匀分布在1 ~ 9号染色体上,平均每个株系携带1.3个染色体片段,平均替换长度为5.62 Mb。替换片段总长度为573.74 Mb,覆盖亲本R4-P1全基因组的73.4%,其中单片段替换片段总长度为139.91 Mb,覆盖全基因组的26.5%,双片段替换片段总长231.41 Mb,覆盖亲本R4-P1全基因组的43.82%,含有3个替换片段总长度为201.3 Mb,覆盖供体亲本基因组25.98%。
中图分类号:
崔建, 钟雄辉, 刘泽慈, 陈登辉, 李海龙, 韩睿, 乐祥庆, 康俊根, 王超. 结球甘蓝染色体片段替换系构建[J]. 园艺学报, 2023, 50(1): 65-78.
CUI Jian, ZHONG Xionghui, LIU Zeci, CHEN Denghui, LI Hailong, HAN Rui, YUE Xiangqing, KANG Jungen, WANG Chao. Construction of Cabbage Chromosome Segment Substitution Lines[J]. Acta Horticulturae Sinica, 2023, 50(1): 65-78.
| 染色体 Chremosome | 标记数 Number of marker | 平均距离/Mb A average distance between 2 markers | 相邻标记最大距离/Mb Maximum distance between adjacent markers |
|---|---|---|---|
| C01 | 16 | 1.76 | 9.69 |
| C02 | 10 | 4.22 | 9.50 |
| C03 | 7 | 4.60 | 16.86 |
| C04 | 15 | 2.45 | 7.66 |
| C05 | 10 | 2.34 | 5.59 |
| C06 | 7 | 5.02 | 10.24 |
| C07 | 23 | 1.20 | 12.58 |
| C08 | 16 | 1.16 | 12.40 |
| C09 | 13 | 2.04 | 18.66 |
| 合计Total | 113 | 2.75 |
表1 各标记在染色体上分布特征
Table 1 Distribution characteristics of each marker on chromosome
| 染色体 Chremosome | 标记数 Number of marker | 平均距离/Mb A average distance between 2 markers | 相邻标记最大距离/Mb Maximum distance between adjacent markers |
|---|---|---|---|
| C01 | 16 | 1.76 | 9.69 |
| C02 | 10 | 4.22 | 9.50 |
| C03 | 7 | 4.60 | 16.86 |
| C04 | 15 | 2.45 | 7.66 |
| C05 | 10 | 2.34 | 5.59 |
| C06 | 7 | 5.02 | 10.24 |
| C07 | 23 | 1.20 | 12.58 |
| C08 | 16 | 1.16 | 12.40 |
| C09 | 13 | 2.04 | 18.66 |
| 合计Total | 113 | 2.75 |
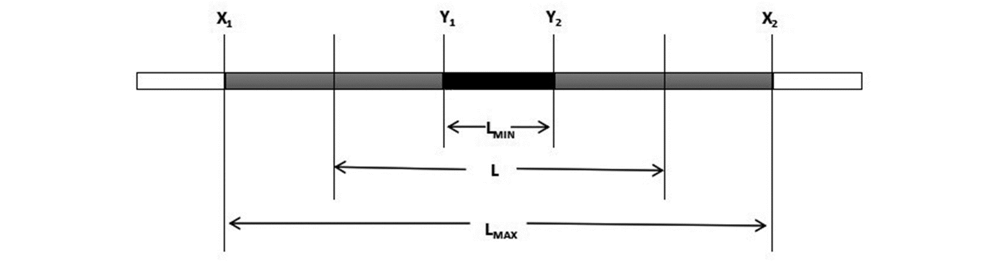
图2 染色体片段替换长度示意图 X代表受体亲代基因型,Y代表供体亲本基因型,L代表染色体片段估计长度,LMAX、LMIN分别代表最大长度和最小长度。
Fig. 2 Schematic diagram of replacement length of chromosome fragments X represents the recipient parental genotype,Y represents the donor parental genotype,L represents the estimated length of the chromosome segment,LMAX and LMIN represent the maximum and minimum lengths.
| 染色体 Chromosome | 代换片段数 Number | 长度范围/Mb Lengh range | 总长度/Mb Total length | 平均长度/Mb Average length | 覆盖长度/Mb Coverage length |
|---|---|---|---|---|---|
| 1 | 12 | 0.45 ~ 9.69 | 41.79 | 3.49 | 41.79 |
| 2 | 11 | 0.24 ~ 18.67 | 101.11 | 9.19 | 38.22 |
| 3 | 4 | 4.61 ~ 9.13 | 24.65 | 6.17 | 19.79 |
| 4 | 14 | 0.27 ~ 13.41 | 59.81 | 4.27 | 34.90 |
| 5 | 7 | 0.80 ~ 5.59 | 17.91 | 2.56 | 17.91 |
| 6 | 8 | 0.26 ~ 26.66 | 95.70 | 11.96 | 32.14 |
| 7 | 17 | 0.06 ~ 14.69 | 54.83 | 3.22 | 38.19 |
| 8 | 12 | 0.13 ~ 12.40 | 42.79 | 3.57 | 38.97 |
| 9 | 17 | 0.49 ~ 46.30 | 135.21 | 7.95 | 49.23 |
| 总计Total | 102 | 0.81 ~ 17.39 | 573.80 | 5.82 | 532.00 |
表2 甘蓝染色体片段替换系遗传评价
Table 2 Genetic evaluation of Chromosome segment substitution lines
| 染色体 Chromosome | 代换片段数 Number | 长度范围/Mb Lengh range | 总长度/Mb Total length | 平均长度/Mb Average length | 覆盖长度/Mb Coverage length |
|---|---|---|---|---|---|
| 1 | 12 | 0.45 ~ 9.69 | 41.79 | 3.49 | 41.79 |
| 2 | 11 | 0.24 ~ 18.67 | 101.11 | 9.19 | 38.22 |
| 3 | 4 | 4.61 ~ 9.13 | 24.65 | 6.17 | 19.79 |
| 4 | 14 | 0.27 ~ 13.41 | 59.81 | 4.27 | 34.90 |
| 5 | 7 | 0.80 ~ 5.59 | 17.91 | 2.56 | 17.91 |
| 6 | 8 | 0.26 ~ 26.66 | 95.70 | 11.96 | 32.14 |
| 7 | 17 | 0.06 ~ 14.69 | 54.83 | 3.22 | 38.19 |
| 8 | 12 | 0.13 ~ 12.40 | 42.79 | 3.57 | 38.97 |
| 9 | 17 | 0.49 ~ 46.30 | 135.21 | 7.95 | 49.23 |
| 总计Total | 102 | 0.81 ~ 17.39 | 573.80 | 5.82 | 532.00 |
| 相关性状 Correlated character | 基因或QTL Gene or QTL | 连锁标记 Linked marker | 参考文献 Reference | 染色体物理位置 Physical position in chromosome | 群体中的位置 Closest markers in the genetic map | 替换片段 长/Mb Length of replacement segment | 连锁单株号 Serial number of linkage line |
|---|---|---|---|---|---|---|---|
| 抗根肿病 Tolerance clubro | Pb(Anju)1a | KBrH059L13 | Nagaoka et al., | C02:24842317 ~ 24842620 | 8C022 ~ BoSF2745 | 7.98 | 29 |
| 抗黑腐病 Resistance to black rot | BRQTL-C1-1a Bo1g056920b | BbRSdcaps-11 | Lee et al., | C01:14884502 ~ 16579946 C01:25091903 ~ 25095843 | BoSF1426 ~ BoSF2345 BoSF1245 ~ BoABL1 | 6.74 9.69 | 5 |
| QTL-1a | BoCL5989s | Kifuji et al., | C02 | 8C022 ~ BoSF2745 | 7.98 | 29 | |
| 枯萎病 Blight | FOCa re-Bol037156b | V17 ~ S9 | Lv et al., | C06:40618342 ~ 40705720 C06 | 8C0464 ~ BoE569 | 5.21 | 95 |
| 抽薹 Bolting | FLCb | — | Lin et al., | C03:2116755 ~ 2137112 | S31 ~ CG750001 | 4.86 | 41 |
| FLC1a | PC14 ~ AC-CATR14 | Razi et al., | C09 | BoSF2645 ~ CK610011 | 18.66 | 163 | |
| COa | PO118E2~PO160E1 | Bohuon et al., | C09 | BoSF2645 ~ CK610011 | 18.66 | 163 | |
| 耐裂球 Crack- resistant ball | SPL-2-2a SPL-4-1a | BRPGM06 BRMS111 | Pang et al., | C02 C04 | 8C022 ~ BoSF2745 CD834770 ~ BoSF1269 | 7.98 5.75 | — 65 |
表3 部分已报道的基因或QTL位点在本群体中的位置
Table 3 The position of some reported genes or QTL loci in this study
| 相关性状 Correlated character | 基因或QTL Gene or QTL | 连锁标记 Linked marker | 参考文献 Reference | 染色体物理位置 Physical position in chromosome | 群体中的位置 Closest markers in the genetic map | 替换片段 长/Mb Length of replacement segment | 连锁单株号 Serial number of linkage line |
|---|---|---|---|---|---|---|---|
| 抗根肿病 Tolerance clubro | Pb(Anju)1a | KBrH059L13 | Nagaoka et al., | C02:24842317 ~ 24842620 | 8C022 ~ BoSF2745 | 7.98 | 29 |
| 抗黑腐病 Resistance to black rot | BRQTL-C1-1a Bo1g056920b | BbRSdcaps-11 | Lee et al., | C01:14884502 ~ 16579946 C01:25091903 ~ 25095843 | BoSF1426 ~ BoSF2345 BoSF1245 ~ BoABL1 | 6.74 9.69 | 5 |
| QTL-1a | BoCL5989s | Kifuji et al., | C02 | 8C022 ~ BoSF2745 | 7.98 | 29 | |
| 枯萎病 Blight | FOCa re-Bol037156b | V17 ~ S9 | Lv et al., | C06:40618342 ~ 40705720 C06 | 8C0464 ~ BoE569 | 5.21 | 95 |
| 抽薹 Bolting | FLCb | — | Lin et al., | C03:2116755 ~ 2137112 | S31 ~ CG750001 | 4.86 | 41 |
| FLC1a | PC14 ~ AC-CATR14 | Razi et al., | C09 | BoSF2645 ~ CK610011 | 18.66 | 163 | |
| COa | PO118E2~PO160E1 | Bohuon et al., | C09 | BoSF2645 ~ CK610011 | 18.66 | 163 | |
| 耐裂球 Crack- resistant ball | SPL-2-2a SPL-4-1a | BRPGM06 BRMS111 | Pang et al., | C02 C04 | 8C022 ~ BoSF2745 CD834770 ~ BoSF1269 | 7.98 5.75 | — 65 |
| [1] | Ando T, Yamamoto T, Shimizu T, Ma X F, Shomura A, Takeuchi Y, Lin S Y, Yano M. 2008. Genetic dissection and pyramiding of quantitative traits for panicle architecture by using chromosomal segment substitution lines in rice. Theoretical & Applied Genetics, 116 (6):881-890. |
| [2] | Arbelaez J D, Moreno L T, Singh N,Tung, Chih Wei, Maron L G, Ospina Y, Martinez C P, Grenier C, Lorieux M, Ccouch S M. 2015. Development and GBS-genotyping of introgression lines(ILs)using two wild species of rice,O. meridionalis and O. rufipogon,in a common recurrent parent,O. sativa cv. Curinga. Molecular Breeding, 35 (2):81. |
| [3] |
Bohuon J E, Ramsay D L, Craft A J, Arthur E A., Marshall F D. 1998. The association of flowering time quantitative trait loci with duplicated regions and candidate. Genetics, 150:393-401.
doi: 10.1093/genetics/150.1.393 pmid: 9725855 |
| [4] |
Bruce A B. 1910. The mendelian theory of heredity and the augmentation of vigor. Science, 32 (827):627-628.
doi: 10.1126/science.32.827.627-a pmid: 17816706 |
| [5] | Chen Denghui, Li Hailong, Tian Duocheng, Sun Chao, Jie Jianming, Kang Jungeng. 2018. Construction of high genetic linkage map and QTL mapping analysis on bolting in cabbage. Journal of Gansu Agricultural University, 53 (5):21-28. (in Chinese) |
| 陈登辉, 李海龙, 田多成, 孙超, 颉建明, 康俊根. 2018. 结球甘蓝高密度遗传图谱构建及抽薹时间相关QTL定位. 甘肃农业大学学报, 53 (5):21-28. | |
| [6] | Chen Shu-xia, Wang Xiao-wu, Fang Zhi-yuan, Cheng Zhi-hui, Sun Pei-tian. 2003. PAPD-based analysis for the Related to bolting time traits of F2 population from Brassica oleracea var. capitata × B. oleracea var. alboglabra. Acta Hortiulturae Sinica, 30 (4):421-426. (in Chinese) |
| 陈书霞, 王晓武, 方智远, 程智慧, 孙培田. 2003. 芥蓝 × 甘蓝的F2群体抽薹期性状QTL的RAPD标记. 园艺学报, 30 (4):421-426. | |
| [7] | Divya B, Malathi S, Sukumar M, Sarla N. 2019. Development and use of chromosome segment substitution lines as a genetic resource for crop improvement. TAG. Theoretical and applied genetics. Theoretische und Angewandte Genetik, 132 (26):1-25. |
| [8] |
East E M. 1936. Heterosis. Genetics, 21 (4):375-397.
doi: 10.1093/genetics/21.4.375 pmid: 17246801 |
| [9] |
Emanuelli F, Lorenzi S, Grzeskowiak L, Catalano V, Grando M S. 2013. Genetic diversity and population structure assessed by SSR and SNP markers in a large germplasm collection of grape. BMC Plant Biology, 13:39.
doi: 10.1186/1471-2229-13-39 pmid: 23497049 |
| [10] |
Eshed Y, Zamir D. 1995. An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics, 141 (3):1147-1162.
doi: 10.1093/genetics/141.3.1147 pmid: 8582620 |
| [11] | Fonceka D, Tossim H A, Rivallan R, Vignes H, Lacut E, de Bellis F, Faye I, Ndoye O, Leal-Bertioli S C M, Valls J F M, Bertioli D J, Glaszmann J C, Courtois B, Rami J F. 2012. Construction of chromosome segment substitution lines in peanut(Arachis hypogaea L.)using a wild synthetic and QTL mapping for plant morphology. PLoS ONE, 7 (11):AR e48642. |
| [12] |
Ghesquière A, Séquier J, Second G, Lorieux M. 1997. First steps towards a rational use of African rice Oryza glaberrima,in rice breeding through a contig line’concept. Euphytica, 96 (1):31-39.
doi: 10.1023/A:1003045518236 URL |
| [13] |
Guo S B, Wei Y, Li X Q, Liu K Q, Huang F K, Chen C H, Gao G Q. 2013. Development and identification of introgression lines from cross of Oryza sativa and Oryza minuta. Rice Science, 20 (2):95-102.
doi: 10.1016/S1672-6308(13)60111-0 URL |
| [14] |
Gupta M, Mason A S, Batley J, Bharti S, Banga S S. 2016. Molecular-cytogenetic characterization of C-genome chromosome substitution lines in Brassica juncea(L.)Czern and Coss. Theoretical and Applied Genetics, 129 (6):1153-1166.
doi: 10.1007/s00122-016-2692-4 URL |
| [15] |
Howell P M, Lydiate D J, Marshall D F. 1996. Towards developing intervarietal substitution lines in Brassica napus using marker-assisted selection. Genome, 39 (2):348-358.
doi: 10.1139/g96-045 pmid: 18469898 |
| [16] | Huang Jing, Cheng Ming-xing, Tang Ming-qiang, Zhang Feng-qi, Zhang Yuan-yuan, Tong Chao-bo, Liu Yue-ying, Cheng Xiao-hui, Dong Cai-hua, Huang Jun-yan, Liu Sheng-yi. 2016. Genome-wide association study of plant height in rapeseed RIL population. Oil Crop Science, 38 (5):543-548. (in Chinese) |
| 黄静, 程明星, 唐敏强, 张凤启, 张园园, 童超波, 刘越英, 程晓晖, 董彩华, 黄军艳, 刘胜毅. 2016. 甘蓝型油菜RIL群体株高性状的全基因组关联分析. 中国油料作物学报, 38 (5):543-548. | |
| [17] |
Kifuji Y, Hanzawa H, Terasawa Y, Ashutosh, Nishio T. 2013. QTL analysis of black rot resistance in cabbage using newly developed EST-SNP markers. Euphytica, 190 (2):289-295.
doi: 10.1007/s10681-012-0847-1 URL |
| [18] | Kubo T, Aida Y, Nakamura K, Tsunematsu H, Doi K, Yoshimura A. 2002. Reciprocal chromosome segment substitution series derived from Japonica and Indica cross of rice(Oryza sativa L.). Breeding Science, 52 (4):319-325. |
| [19] |
Lee J, Izzah N, Jayakodi M, Perumal S, Joh H, Lee H, Lee S C, Park J, Yang K W, Nou I S. 2015. Genome-wide SNP identification and QTL mapping for black rot resistance in cabbage. Bmc Plant Biology, 15:32.
doi: 10.1186/s12870-015-0424-6 pmid: 25644124 |
| [20] | Li X, Wang W, Zhe W, Li K, Pyo L Y, Zhongyun P. 2015. Construction of chromosome segment substitution lines enables QTL mapping for flowering and morphological traits in Brassica rapa. Frontiers in Plant Science, 1 (6):432. |
| [21] | Li Xuefeng, Hu Xiaowen, Zhang Shengping, Gu Xingfang, Zhang Zhonghua, Huang Sanwen. 2011. Construction of wild cucumber substitution lines. Acta Hortiulturae Sinica, 38 (5):886-892. (in Chinese) |
| 李学峰, 胡晓文, 张圣平, 顾兴芳, 张忠华, 黄三文. 2011. 野生黄瓜代换系的构建. 园艺学报, 38 (5):886-892. | |
| [22] |
Lin S I, Wang J G, Poon S Y, Su C L, Wang S S, Chiou T J. 2005. Differential regulation of FLOWERING LOCUS C expression by vernalization in cabbage and Arabidopsis. Plant Physiology, 137 (3):1037-1048.
doi: 10.1104/pp.104.058974 URL |
| [23] | Liu S, Zhou R, Dong Y, Pei L, Jia J. 2006. Development,utilization of introgression lines using a synthetic wheat as donor. Theoretical & Applied Genetics, 112 (7):1360-1373. |
| [24] | Liu Zeci. 2019. Construction of cabbage(Brassica oleracea var. capitata)Chromosome segment substitution lines and black rot resistance mapping and candidate genes analysis [Ph. D. Dissertation]. Lanzhou: Gansu Agricultural University. (in Chinese) |
| 刘泽慈. 2019. 甘蓝染色体片段替换系的构建和黑腐病抗性QTL定位及候选基因分析[博士论文]. 兰州: 甘肃农业大学. | |
| [25] |
Luan M, Guo X, Zhang Y, Yao J, Chen W. 2010. QTL mapping for agronomic and fibre traits using two interspecific chromosome substitution lines of upland cotton. Plant Breeding, 128 (6):671-679.
doi: 10.1111/j.1439-0523.2009.01650.x URL |
| [26] |
Lv H, Fang Z, Yang L, Zhang Y, Wang Q, Liu Y, Zhuang M, Yang Y, Xie B, Liu B. 2014. Mapping and analysis of a novel candidate Fusarium wilt resistance gene FOC1 in Brassica oleracea. BMC Genomics, 15 (1):1094.
doi: 10.1186/1471-2164-15-1094 URL |
| [27] |
McCouch S R, Sweeney M, Li J, Jiang H, Thomson M, Septiningsih E. 2007. Through the genetic bottleneck:O. rufipogon as a source of trait-enhancing alleles for O. sativa. Euphytica, 154:317-39.
doi: 10.1007/s10681-006-9210-8 URL |
| [28] |
Nagaoka T, Doullah M, Matsumoto S, Kawasaki S, Ishikawa T, Hori H, Okazaki K. 2010. Identification of QTLs that control clubroot resistance in Brassica oleracea and comparative analysis of clubroot resistance genes between B. rapa and B. oleracea. Theoretical and Applied Genetics, 120 (7):1335-1346.
doi: 10.1007/s00122-010-1259-z pmid: 20069415 |
| [29] | Okazaki K, Sakamoto K, Kikuchi R, Saito A, Togashi E, Kuginuki Y, Matsumoto S, Hirai M. 2007. Mapping and characterization of FLC homologs and QTL analysis of flowering time in Brassica oleracea. Theoretical & Applied Genetics, 114 (4):595-608. |
| [30] |
Pang W, Li X, Su R C, Nguyen V D, Yong P L Dhandapani V, Kim Y Y, Ramchiary N, Kim J G, Edwards D, Batley J, Na J, Kim H. 2015. Mapping QTLs of resistance to head splitting in cabbage(Brassica oleracea L. var. capitata L.). Molecular Breeding, 35 (5):1-12.
doi: 10.1007/s11032-015-0202-z URL |
| [31] | Peng X, He H, Zhu G, Jiang L, Zhu C, Yu Q, He J, Shen X, Yan S, Bian J. 2017. Identification and analyses of chromosome segments affecting heterosis using chromosome-segment substitution lines in rice. Crop Ence, 57 (4):1836-1843. |
| [32] |
Ramsay L D, Jennings D E, Bohuon E J R, Arthur A E, Lydiate D J, Kearsey M J, Marshall D F. 1996. The construction of a substitution library of recombinant backcross lines in Brasscia oleracea for the precision mapping of quantitative trait loci. Genome, 39:558-567.
doi: 10.1139/g96-071 pmid: 18469917 |
| [33] |
Razi H, Howell E C, Newbury H J, Kearsey M J. 2008. Does sequence polymorphism of FLC paralogues underlie flowering time QTL in Brassica oleracea? Theoretical and Applied Genetics, 116 (2):179-192.
doi: 10.1007/s00122-007-0657-3 pmid: 17938878 |
| [34] | Su Yanbin, Li Qiang, Yi Dengxia, Liu Lujiang, Fu Changzhi, Zhang Tianzhu, Zhang Haizhen, Chen Yanhong. 2019. Genetic analysis of traits related to head in white cabbage. Northern Horticulture,(8):7-14. (in Chinese) |
| 苏彦宾, 李强, 仪登霞, 刘鲁江, 傅长智, 张天柱, 张海珍, 陈燕红. 2019. 结球甘蓝叶球相关性状遗传分析. 北方园艺,(8):7-14. | |
| [35] | Szalma S J, Hostert B M, Ledeaux J R, Stuber C W, Holland J B. 2007. QTL mapping with near-isogenic lines in maize. Theoretical & Applied Genetics, 114 (7):1211-1228. |
| [36] | Tian Duo-cheng. 2014. Construcion of a molecular high-density genetic map and mapping of QTLs related to glucosinolated traits in cabbage (Brassica oleracea var. capitata)[Ph. D. Dissertation]. Gansu: Gansu Agricultural University. (in Chinese) |
| 田多成. 2014. 结球甘蓝高密度分子遗传图谱的构建及硫代葡萄糖苷相关性状的QTL定位分析[博士论文]. 兰州: 甘肃农业大学. | |
| [37] |
Wan X Y, Wan J M, Jiang L, Wang J K, Zhai H Q, Weng J F. 2006. QTL analysis for rice grain length and fine mapping of an identified QTL with stable and major effects. Theor Appl Genet, 112:1258-1270.
doi: 10.1007/s00122-006-0227-0 pmid: 16477428 |
| [38] | Wang Liqiu, Zhao Yongfeng, Xue Yadong, Zhang Zuxin, Zhen Yonglian, Chen Jingtang. 2007. Development and evaluation of two link-up single segment introgression lines(SSILs)in Zea mays. Acta Agronomica Sinica,(4):663-668. (in Chinese) |
| 王立秋, 赵永锋, 薛亚东, 张祖新, 郑用琏, 陈景堂. 2007. 玉米衔接式单片段导入系群体的构建和评价. 作物学报,(4):663-668. | |
| [39] | Wang Wuhong, Wang Jinglei, li Biyuan, Wei Qingzhen, Hu Tianhua, Hu Haijiao, Bao Chonglai. 2020. Genetic and QTL mapping analysis of bolting time in cabbsge(Brassica oleracea). Acta Hortiulturae Sinica, 47 (5):974-982. (in Chinese) |
| 王五宏, 汪精磊, 李必元, 魏庆镇, 胡天华, 胡海娇, 包崇来. 2020. 结球甘蓝抽薹性遗传规律和QTL定位分析. 园艺学报, 47 (5):974-982. | |
| [40] |
Wang Y, Wang X, Wang X, Zhao Q, Lv X, Feng H. 2018. Construction of chromosome segment substitution lines of Chinese cabbage(Brassica rapa L. ssp. pekinensis)in the background of RcBr(B. rapa L. ssp. dichotoma)and characterization of segments representing the bolting trait. Molecular Breeding, 38 (4):35.
doi: 10.1007/s11032-018-0794-1 URL |
| [41] |
Wang Y F, Zhang X G, Shi X, Sun C, Jiao J, Tian R M, Wei X Y, Xie H L, Guo Z Y, Tang J H. 2018. Heterotic loci identified for maize kernel traits in two chromosome segment substitution line test populations. Scientific Reports, 8 (1):11101.
doi: 10.1038/s41598-018-29338-1 pmid: 30038303 |
| [42] | Wang Yunpeng, Wang Xingfen, Li Zhikun, Yang Xinlei, Zhang Yan, Wu Liqiang, Wu Jinhua, Zhang Guiyin, Ma Zhiying. 2016. Development of Pima cotton chromosome segment substitution lines with Gossypium hirsutum background. Journal of Plant Genetic Resources, 17 (1):114-119. (in Chinese) |
| 王云鹏, 王省芬, 李志坤, 杨鑫雷, 张艳, 吴立强, 吴金华, 张桂寅, 马峙英. 2016. 陆地棉背景的Pima棉染色体片段置换系创制. 植物遗传资源学报, 17 (1):114-119. | |
| [43] | Wei H, Jin J, Sun S Y, Zhu M Z. 2006. Construction of chromosome segment substitution lines carrying overlapping chromosome segments of the whole wild rice genome and identification of quantitative trait loci for rice quality. Journal of Plant Physiology & Molecular Biology, 32 (3):354. |
| [44] |
William R L, Pollak E. 1985. Theory of heterosis. Journal of Dairy Science, 68 (9):2411-2417.
doi: 10.3168/jds.S0022-0302(85)81117-6 URL |
| [45] |
Xi Z Y, He F H, Zeng R Z, Zhang Z M, Ding X H, Li W T, Zhang G Q. 2006. Development of a wide population of chromosome single-segment substitution lines in the genetic background of an elite cultivar of rice(Oryza sativa L.). Genome, 49 (5):476-484.
doi: 10.1139/g06-005 pmid: 16767172 |
| [46] | Xu Rui, Zhang Lizhen, Xie Xianzhi, Wang Yanyan, Huang Jingfen, Zheng Xiaoming, Zhang Lifang, Cheng Yunlian, Zheng Chongke, Qiao Weihua, Yang Qingwen. 2020. QTL mapping revealed salt tolerance traits from Oryza rufipogon using chromosome segment substitution lines. Journal of Plant Genetic Resources, 21 (2):442-451. (in Chinese) |
| 许睿, 张莉珍, 谢先芝, 王艳艳, 黄婧芬, 郑晓明, 张丽芳, 程云连, 郑崇珂, 乔卫华, 杨庆文. 2020. 基于染色体置换系的普通野生稻耐盐性QTL定位. 物遗传资源学报, 21 (2):442-451. | |
| [47] | Yang Limei, Fang Zhiyuan. 2022. Researches on cabbage genetics and breeding in China for 60 years. Acta Horticulturae Sinica, 49 (10):2075-2098. (in Chinese) |
| 杨丽梅, 方智远. 2022. 中国甘蓝遗传育种研究60年. 园艺学报, 49 (10):2075-2098. | |
| [48] | Young N D, Tanksley S D. 1989. Restriction fragment length polymorphism maps and the concept of graphical genotypes. Theoretical & Applied Genetics, 77 (1):95-101. |
| [49] |
Yu C, Wan J, Zhai H, Wang C, Liu Y. 2005. Study on heterosis of inter-subspecies between indica and japonica rice(Oryza sativa L.)using chromosome segment substitution lines. Chinese Science Bulletin, 50 (2):131-136.
doi: 10.1007/BF02897516 URL |
| [50] | Zhu Xiaowei, Bo Tianyue, Chen Jinxiu, Tai Xiang, Ren Yunying. 2017. Research progress on mapping of genes associated with main agronomic traits of cabbage. Acta Hortiulturae Sinica, 44 (9):1729-1737. (in Chinese) |
| 朱晓炜, 薄天岳, 陈锦秀, 邰翔, 任云英. 2017. 结球甘蓝主要农艺性状基因定位的研究进展. 园艺学报, 44 (9):1729-1737. |
| [1] | 刘艺平, 倪梦辉, 吴芳芳, 刘红利, 贺丹, 孔德政. 荷花花器官性状与SSR标记的关联分析[J]. 园艺学报, 2023, 50(1): 103-115. |
| [2] | 罗海林, 袁雷, 翁华, 闫佳会, 郭青云, 王文清, 马新明. 蚕豆萎蔫病毒2号青海辣椒分离物的鉴定与全基因组序列克隆[J]. 园艺学报, 2023, 50(1): 161-169. |
| [3] | 吕红豪, 杨丽梅, 方智远, 张扬勇, 庄 木, 刘玉梅, 王 勇, 季家磊, 李占省, 韩风庆. 春甘蓝新品种‘中甘27’和‘中甘28’[J]. 园艺学报, 2022, 49(S1): 63-64. |
| [4] | 王惠哲, 杨瑞环, 邓 强, 曹明明, 李淑菊, . 抗黑星病黄瓜新品种‘津冬369’[J]. 园艺学报, 2022, 49(S1): 85-86. |
| [5] | 杨兴玉, 许林兵, 吴元立, 邱迪洋, 范琳琳, 黄秉智. 香蕉杂交种‘粉杂1号’ABBB基因组鉴定[J]. 园艺学报, 2022, 49(9): 1991-1997. |
| [6] | 张鲁刚, 卢倩倩, 何琼, 薛一花, 马晓敏, 马帅, 聂姗姗, 杨文静. 紫橙色大白菜新种质的创制[J]. 园艺学报, 2022, 49(7): 1582-1588. |
| [7] | 肖学宸, 刘梦雨, 蒋梦琦, 陈燕, 薛晓东, 周承哲, 吴兴健, 吴君楠, 郭寅生, 叶开温, 赖钟雄, 林玉玲. 龙眼褪黑素合成途径SNAT、ASMT和COMT家族基因鉴定及表达分析[J]. 园艺学报, 2022, 49(5): 1031-1046. |
| [8] | 潘鑫峰, 叶方婷, 毛志君, 李兆伟, 范凯. 睡莲WRKY家族的全基因组鉴定和分子进化分析[J]. 园艺学报, 2022, 49(5): 1121-1135. |
| [9] | 李莎莎, 玉赛赛, 傅雨恒, 骆强伟, 徐炎, 王跃进. 利用胚挽救与分子标记选育葡萄无核抗寒新种质[J]. 园艺学报, 2022, 49(4): 723-738. |
| [10] | 周陈平, 杨敏, 郭金菊, 邝瑞彬, 杨护, 黄炳雄, 魏岳荣. 番木瓜成熟过程中全基因组DNA甲基化和转录组变化分析[J]. 园艺学报, 2022, 49(3): 519-532. |
| [11] | 刘梦雨, 蒋梦琦, 陈燕, 张舒婷, 薛晓东, 肖学宸, 赖钟雄, 林玉玲. 龙眼GDSL酯酶/脂肪酶基因的全基因组鉴定及表达分析[J]. 园艺学报, 2022, 49(3): 597-612. |
| [12] | 乔军, 刘婧, 李素文, 王利英. 基于极端混合池全基因组重测序的茄子萼下果色基因预测[J]. 园艺学报, 2022, 49(3): 613-621. |
| [13] | 卢甜甜, 刘志远, 徐兆生, 张合龙, 李国亮, 折红兵, 钱伟. 菜豆花色全基因组关联分析[J]. 园艺学报, 2022, 49(2): 332-340. |
| [14] | 方天, 刘继红. 主要果树重要性状QTL鉴定研究进展[J]. 园艺学报, 2022, 49(12): 2622-2640. |
| [15] | 宋蒙飞, 查高辉, 陈劲枫, 娄群峰. 黄瓜株型性状分子基础研究进展[J]. 园艺学报, 2022, 49(12): 2683-2702. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司