
园艺学报 ›› 2022, Vol. 49 ›› Issue (11): 2313-2324.doi: 10.16420/j.issn.0513-353x.2021-0863
聂兴华1,2, 李伊然2, 田寿乐3, 王雪峰4, 苏淑钗1, 曹庆芹2, 邢宇2,*( ), 秦岭2,*(
), 秦岭2,*( )
)
收稿日期:2022-04-26
修回日期:2022-07-28
出版日期:2022-11-25
发布日期:2022-11-25
通讯作者:
邢宇,秦岭
E-mail:xingyu@bua.edu.cn;qinlingbac@126.com
基金资助:
NIE Xinghua1,2, LI Yiran2, TIAN Shoule3, WANG Xuefeng4, SU Shuchai1, CAO Qingqin2, XING Yu2,*( ), QIN Ling2,*(
), QIN Ling2,*( )
)
Received:2022-04-26
Revised:2022-07-28
Online:2022-11-25
Published:2022-11-25
Contact:
XING Yu,QIN Ling
E-mail:xingyu@bua.edu.cn;qinlingbac@126.com
摘要:
基于SSR荧光分子标记,对中国342个板栗品种(系)构建了DNA指纹图谱,并解析了各品种群间的亲缘关系和遗传分化特征。结果表明:(1)所有SSR标记多位点匹配后的PI和PIsibs值分别为4.960 × 10-20 和2.395 × 10-8,显示本试验所选用的标记指纹识别潜力强;对342份品种(系)构建指纹图谱,共获得338个有唯一对应关系的指纹信息,其中舒城大红袍与焦扎、双合大红袍与CSB-1两两共享了一个指纹信息,结合主要植物学性状,推测其为同物异名品种;同时发现存在大于10个位点信息差异的19份同名资源,判定其为同名异物品种。(2)两两品种群的Fst平均值为0.0224(Fst < 0.05),表明品种群间遗传分化程度较低;对5个品种群进行了基因流(Nm)和AMOVA分析,遗传变异主要存在于个体内,占总变异的84%,各品种群间的平均基因流为6.592,表明资源间存在着广泛的基因交换,制约了各群体间的分化。(3)利用位点数据进行群体结构、主坐标和聚类树分析表明,板栗品种资源主要被划分为南、北两大品种群,确定了沿大别山山脉的湖北东北部、河南南部和安徽中北部、以及山东南部和江苏北部地区为分界带(混交地带)。
中图分类号:
聂兴华, 李伊然, 田寿乐, 王雪峰, 苏淑钗, 曹庆芹, 邢宇, 秦岭. 中国板栗品种(系)DNA指纹图谱构建及其遗传多样性分析[J]. 园艺学报, 2022, 49(11): 2313-2324.
NIE Xinghua, LI Yiran, TIAN Shoule, WANG Xuefeng, SU Shuchai, CAO Qingqin, XING Yu, QIN Ling. Construction of DNA Fingerprint Map and Analysis of Genetic Diversity for Chinese Chestnut Cultivars(Lines)[J]. Acta Horticulturae Sinica, 2022, 49(11): 2313-2324.
| 编号 Number | 分子标记 Marker ID | 基因库ID GenBank ID | 正向引物序列(5′-3′) Forward primer | 反向引物序列(5′-3′) Reverse primer | 观察的重复序列 Observed-Motif |
|---|---|---|---|---|---|
| 1 | CmSI0396 | 290474606 | AACTCCCACCACTCACATCC | TTTCGGACCATCCAGAACTC | CACACC |
| 2 | CmSI0509 | 290474647 | CAAGTCCGATCCTTCCTCTG | AGCTGGGTTTTGAGTAGCGA | ACA |
| 3 | CmSI0561 | 290474702 | CGTATAGGGTGGAAACGGAA | GGACAAGCAAATCACGGAAT | TCG |
| 4 | CmSI0614 | 290476556 | TTGTGGTGAAGCTGACATCG | GGGTACTACCACAACATGCAG | GTT |
| 5 | CmSI0617 | 290475843 | GCCTCAGCTGGTAACGAAGA | TCACATGGGCTTGTTGTAGC | GA |
| 6 | CmSI0658 | 290474781 | AAAACGGTTTGTGGTGAAGC | GCCAACCAGTCAAGGGTACT | GTT |
| 7 | CmSI0702 | 290474818 | GAAACACACCAGAGAGATGCAG | TTTTATACAGAGACATACTATCCTACACAG | TC |
| 8 | CmSI0735 | 290474842 | ACGCCTTCAGTTGCTGTTTC | CAACGGTCTTACCCTTTGGA | AGAA |
| 9 | CmSI0742 | 290474850 | GACGCTCCTCAGCTTTTGAC | TGCCGGTCAATTCTTCTTCT | AG |
| 10 | CmSI0800 | 290474899 | TTATGGCAACCCTCCTGTTT | CTGAAATGATCGATGCTGCT | TC |
| 11 | CmSI0853 | 290474950 | GGAGGAGGAGGAGCTCATTG | CCTTGGAGAGCTGCCAGTAG | TCT |
| 12 | CmSI0871 | 290474967 | AGGGGGTGGAAGAACCTATG | AGATTGCAAGTGGGGAATTG | TCT |
| 13 | CmSI0881 | 290476058 | TCCGACAAGCTACCGAGTCT | CCTAAATCAATCTTGCACCCATA | GA |
| 14 | CmSI0883 | 290476060 | CAGCATCAGCACTCGTTCA | GGGATTGAGAGGATGAAGCA | AGC |
| 15 | CmSI0922 | 290475011 | AATCTGAACCCCTCCGATCT | ACCAACAACATGTGCCAAAA | TTG |
| 16 | CmSI0930 | 290475019 | CCATTTAGCATGCATAGTCATACC | GCAAGGATGTAGGTCGAATCA | ATAC |
| 17 | CmSI0938 | 290475031 | CACGATTCCCACGATTCTTA | CGTCCAACTCCGTACTCTCC | GAC |
| 18 | CmSI0404 | 290474610 | GTGCTACGACTACCGCTGCT | AGAATAACCTCGCGGTGAGA | ACA |
| 19 | CmSI0747 | 290474854 | CATTCCAACAATGCACTCTCA | CGAGTTGGAGTCACCGAAGT | CTT |
| 20 | CmSI0809 | 290474901 | GAGCGAGTCGAAGAAGGAAG | ATCTGCTTCGGCACCATCT | GGT |
| 21 | CmSI0933 | 290475025 | AGGCCTCTTCTCCCTTGTGT | TCCTGTTCCTATGCTGCTCA | AGA |
表1 21对SSR引物的信息
Table 1 The information of 21 pairs of SSR primers
| 编号 Number | 分子标记 Marker ID | 基因库ID GenBank ID | 正向引物序列(5′-3′) Forward primer | 反向引物序列(5′-3′) Reverse primer | 观察的重复序列 Observed-Motif |
|---|---|---|---|---|---|
| 1 | CmSI0396 | 290474606 | AACTCCCACCACTCACATCC | TTTCGGACCATCCAGAACTC | CACACC |
| 2 | CmSI0509 | 290474647 | CAAGTCCGATCCTTCCTCTG | AGCTGGGTTTTGAGTAGCGA | ACA |
| 3 | CmSI0561 | 290474702 | CGTATAGGGTGGAAACGGAA | GGACAAGCAAATCACGGAAT | TCG |
| 4 | CmSI0614 | 290476556 | TTGTGGTGAAGCTGACATCG | GGGTACTACCACAACATGCAG | GTT |
| 5 | CmSI0617 | 290475843 | GCCTCAGCTGGTAACGAAGA | TCACATGGGCTTGTTGTAGC | GA |
| 6 | CmSI0658 | 290474781 | AAAACGGTTTGTGGTGAAGC | GCCAACCAGTCAAGGGTACT | GTT |
| 7 | CmSI0702 | 290474818 | GAAACACACCAGAGAGATGCAG | TTTTATACAGAGACATACTATCCTACACAG | TC |
| 8 | CmSI0735 | 290474842 | ACGCCTTCAGTTGCTGTTTC | CAACGGTCTTACCCTTTGGA | AGAA |
| 9 | CmSI0742 | 290474850 | GACGCTCCTCAGCTTTTGAC | TGCCGGTCAATTCTTCTTCT | AG |
| 10 | CmSI0800 | 290474899 | TTATGGCAACCCTCCTGTTT | CTGAAATGATCGATGCTGCT | TC |
| 11 | CmSI0853 | 290474950 | GGAGGAGGAGGAGCTCATTG | CCTTGGAGAGCTGCCAGTAG | TCT |
| 12 | CmSI0871 | 290474967 | AGGGGGTGGAAGAACCTATG | AGATTGCAAGTGGGGAATTG | TCT |
| 13 | CmSI0881 | 290476058 | TCCGACAAGCTACCGAGTCT | CCTAAATCAATCTTGCACCCATA | GA |
| 14 | CmSI0883 | 290476060 | CAGCATCAGCACTCGTTCA | GGGATTGAGAGGATGAAGCA | AGC |
| 15 | CmSI0922 | 290475011 | AATCTGAACCCCTCCGATCT | ACCAACAACATGTGCCAAAA | TTG |
| 16 | CmSI0930 | 290475019 | CCATTTAGCATGCATAGTCATACC | GCAAGGATGTAGGTCGAATCA | ATAC |
| 17 | CmSI0938 | 290475031 | CACGATTCCCACGATTCTTA | CGTCCAACTCCGTACTCTCC | GAC |
| 18 | CmSI0404 | 290474610 | GTGCTACGACTACCGCTGCT | AGAATAACCTCGCGGTGAGA | ACA |
| 19 | CmSI0747 | 290474854 | CATTCCAACAATGCACTCTCA | CGAGTTGGAGTCACCGAAGT | CTT |
| 20 | CmSI0809 | 290474901 | GAGCGAGTCGAAGAAGGAAG | ATCTGCTTCGGCACCATCT | GGT |
| 21 | CmSI0933 | 290475025 | AGGCCTCTTCTCCCTTGTGT | TCCTGTTCCTATGCTGCTCA | AGA |
| 分子标记 Marker ID | 主要位点 频率MAF | 基因型数Ng | 位点数 Na | Shannon多样性指数I | 观察杂合度Ho | 期望杂合度He | 基因流 Nm | 多态信息 含量 PIC | 身份概率 PI | 随机身份 概率 PIsibs |
|---|---|---|---|---|---|---|---|---|---|---|
| CmSI0396 | 0.568 | 9 | 4 | 0.989 | 0.589 | 0.567 | 7.678 | 0.492 | 0.261 | 0.532 |
| CmSI0509 | 0.477 | 13 | 6 | 1.179 | 0.542 | 0.646 | 4.987 | 0.583 | 0.188 | 0.474 |
| CmSI0561 | 0.509 | 32 | 14 | 1.602 | 0.630 | 0.694 | 3.793 | 0.673 | 0.118 | 0.433 |
| CmSI0617 | 0.325 | 30 | 9 | 1.713 | 0.714 | 0.778 | 3.482 | 0.744 | 0.081 | 0.381 |
| CmSI0658 | 0.249 | 22 | 7 | 1.703 | 0.752 | 0.802 | 5.099 | 0.774 | 0.069 | 0.366 |
| CmSI0702 | 0.412 | 26 | 11 | 1.409 | 0.647 | 0.702 | 3.202 | 0.655 | 0.140 | 0.434 |
| CmSI0735 | 0.403 | 27 | 10 | 1.604 | 0.557 | 0.732 | 9.646 | 0.697 | 0.109 | 0.411 |
| CmSI0742 | 0.422 | 20 | 13 | 1.346 | 0.627 | 0.689 | 6.857 | 0.633 | 0.151 | 0.443 |
| CmSI0800 | 0.326 | 58 | 22 | 2.020 | 0.633 | 0.802 | 8.051 | 0.777 | 0.063 | 0.365 |
| CmSI0809 | 0.270 | 24 | 10 | 1.824 | 0.624 | 0.809 | 2.930 | 0.762 | 0.063 | 0.362 |
| CmSI0853 | 0.301 | 26 | 14 | 1.740 | 0.749 | 0.792 | 7.355 | 0.762 | 0.073 | 0.372 |
| CmSI0871 | 0.597 | 15 | 11 | 0.920 | 0.522 | 0.527 | 11.653 | 0.448 | 0.306 | 0.563 |
| CmSI0883 | 0.278 | 33 | 13 | 1.787 | 0.636 | 0.798 | 4.404 | 0.770 | 0.070 | 0.368 |
| CmSI0933 | 0.646 | 10 | 7 | 0.936 | 0.493 | 0.514 | 10.042 | 0.459 | 0.294 | 0.567 |
| CmSI0938 | 0.785 | 14 | 7 | 0.755 | 0.204 | 0.364 | 10.284 | 0.332 | 0.432 | 0.676 |
| CmSI0922 | 0.254 | 38 | 12 | 1.944 | 0.738 | 0.833 | 7.656 | 0.811 | 0.049 | 0.346 |
| CmSI0930 | 0.419 | 51 | 20 | 1.885 | 0.697 | 0.763 | 9.199 | 0.741 | 0.080 | 0.389 |
| CmSI0881 | 0.324 | 24 | 8 | 1.760 | 0.493 | 0.798 | 6.470 | 0.756 | 0.068 | 0.368 |
| CmSI0614 | 0.212 | 29 | 10 | 1.857 | 0.761 | 0.826 | 6.203 | 0.800 | 0.054 | 0.351 |
| CmSI0404 | 0.839 | 6 | 4 | 0.781 | 0.224 | 0.388 | 2.285 | 0.254 | 0.400 | 0.656 |
| CmSI0747 | 0.549 | 10 | 5 | 1.382 | 0.487 | 0.676 | 7.158 | 0.578 | 0.143 | 0.448 |
| 平均值Mean | 0.436 | 24.619 | 10.333 | 1.483 | 0.587 | 0.690 | 6.592 | 0.643 | 0.153 | 0.443 |
| 组合 Combined | — | — | — | — | — | — | — | — | 4.960 ×10-20 | 2.395×10-8 |
表2 21个SSR分子标记在342份板栗品种中的关键遗传数据
Table 2 The key genetic statistics of 21 SSR markers in 342 Chinese chestnut accessions
| 分子标记 Marker ID | 主要位点 频率MAF | 基因型数Ng | 位点数 Na | Shannon多样性指数I | 观察杂合度Ho | 期望杂合度He | 基因流 Nm | 多态信息 含量 PIC | 身份概率 PI | 随机身份 概率 PIsibs |
|---|---|---|---|---|---|---|---|---|---|---|
| CmSI0396 | 0.568 | 9 | 4 | 0.989 | 0.589 | 0.567 | 7.678 | 0.492 | 0.261 | 0.532 |
| CmSI0509 | 0.477 | 13 | 6 | 1.179 | 0.542 | 0.646 | 4.987 | 0.583 | 0.188 | 0.474 |
| CmSI0561 | 0.509 | 32 | 14 | 1.602 | 0.630 | 0.694 | 3.793 | 0.673 | 0.118 | 0.433 |
| CmSI0617 | 0.325 | 30 | 9 | 1.713 | 0.714 | 0.778 | 3.482 | 0.744 | 0.081 | 0.381 |
| CmSI0658 | 0.249 | 22 | 7 | 1.703 | 0.752 | 0.802 | 5.099 | 0.774 | 0.069 | 0.366 |
| CmSI0702 | 0.412 | 26 | 11 | 1.409 | 0.647 | 0.702 | 3.202 | 0.655 | 0.140 | 0.434 |
| CmSI0735 | 0.403 | 27 | 10 | 1.604 | 0.557 | 0.732 | 9.646 | 0.697 | 0.109 | 0.411 |
| CmSI0742 | 0.422 | 20 | 13 | 1.346 | 0.627 | 0.689 | 6.857 | 0.633 | 0.151 | 0.443 |
| CmSI0800 | 0.326 | 58 | 22 | 2.020 | 0.633 | 0.802 | 8.051 | 0.777 | 0.063 | 0.365 |
| CmSI0809 | 0.270 | 24 | 10 | 1.824 | 0.624 | 0.809 | 2.930 | 0.762 | 0.063 | 0.362 |
| CmSI0853 | 0.301 | 26 | 14 | 1.740 | 0.749 | 0.792 | 7.355 | 0.762 | 0.073 | 0.372 |
| CmSI0871 | 0.597 | 15 | 11 | 0.920 | 0.522 | 0.527 | 11.653 | 0.448 | 0.306 | 0.563 |
| CmSI0883 | 0.278 | 33 | 13 | 1.787 | 0.636 | 0.798 | 4.404 | 0.770 | 0.070 | 0.368 |
| CmSI0933 | 0.646 | 10 | 7 | 0.936 | 0.493 | 0.514 | 10.042 | 0.459 | 0.294 | 0.567 |
| CmSI0938 | 0.785 | 14 | 7 | 0.755 | 0.204 | 0.364 | 10.284 | 0.332 | 0.432 | 0.676 |
| CmSI0922 | 0.254 | 38 | 12 | 1.944 | 0.738 | 0.833 | 7.656 | 0.811 | 0.049 | 0.346 |
| CmSI0930 | 0.419 | 51 | 20 | 1.885 | 0.697 | 0.763 | 9.199 | 0.741 | 0.080 | 0.389 |
| CmSI0881 | 0.324 | 24 | 8 | 1.760 | 0.493 | 0.798 | 6.470 | 0.756 | 0.068 | 0.368 |
| CmSI0614 | 0.212 | 29 | 10 | 1.857 | 0.761 | 0.826 | 6.203 | 0.800 | 0.054 | 0.351 |
| CmSI0404 | 0.839 | 6 | 4 | 0.781 | 0.224 | 0.388 | 2.285 | 0.254 | 0.400 | 0.656 |
| CmSI0747 | 0.549 | 10 | 5 | 1.382 | 0.487 | 0.676 | 7.158 | 0.578 | 0.143 | 0.448 |
| 平均值Mean | 0.436 | 24.619 | 10.333 | 1.483 | 0.587 | 0.690 | 6.592 | 0.643 | 0.153 | 0.443 |
| 组合 Combined | — | — | — | — | — | — | — | — | 4.960 ×10-20 | 2.395×10-8 |
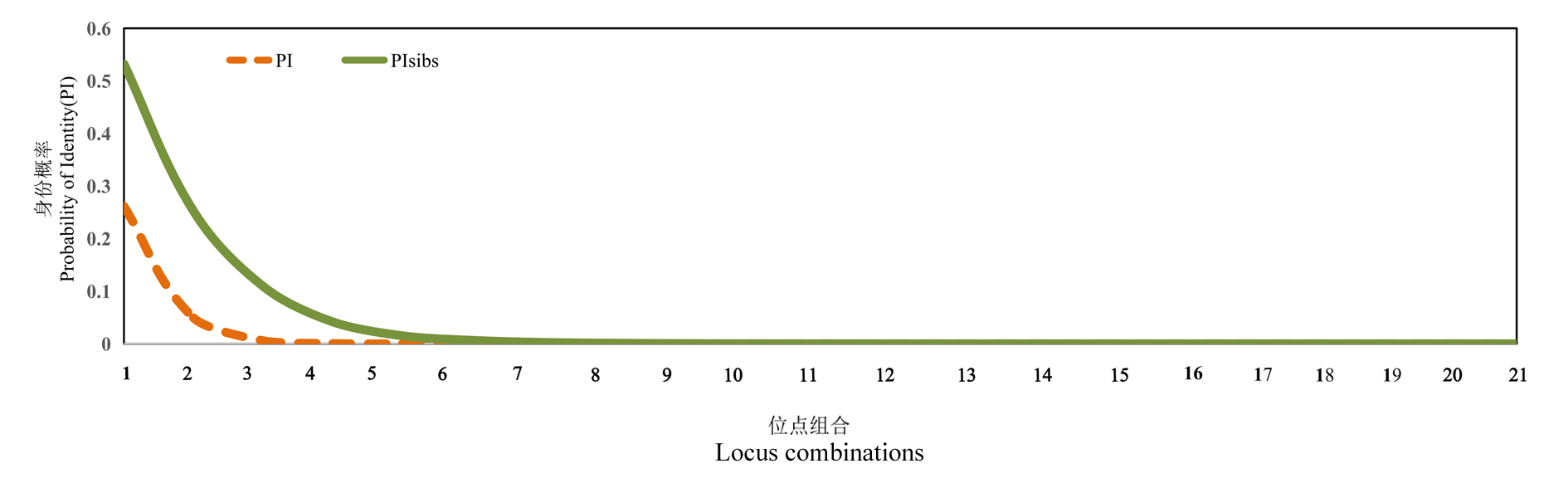
图1 SSR标记在板栗品种指纹鉴定能力中的评价 1代表引物1;2代表引物1 + 引物2;3代表引物1 + 引物2 + 引物3;……;21代表所有21个引物组合。引物序号同表1。
Fig. 1 Evaluation for the fingerprinting power of SSR markers in Chinese chestnut cultivars 1 represents primer 1;2 represents primer 1 + primer 2;3 represents primer 1 + primer 2 + primer 3;......;21 represents all 21 primer combinations. The primer numbers are the same as in Table 1.
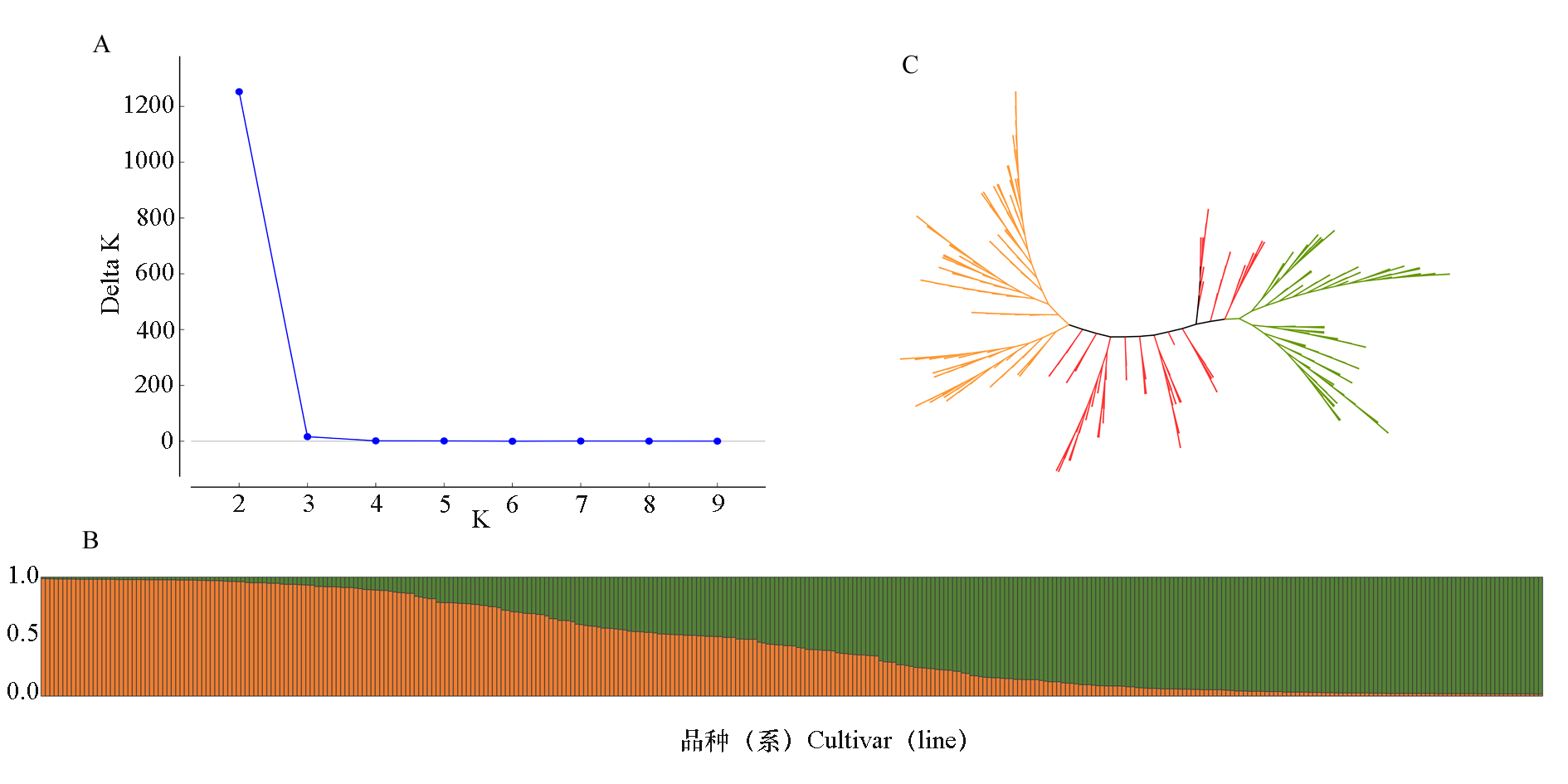
图3 板栗342个品种(系)的群体结构与聚类树 A:群体结构分析的Delta K值分布;B:K = 2时品种群体结构图;C:UPGMA无根树(橙色为group 1,绿色为group 2,红色为group 1和 group 2间的过渡类型)。
Fig. 3 Population structure and cluster tree of 342 Chinese chestnut cultivars(lines) A:Delta K value distribution of population structure analysis;B:Population structure at K = 2;C:UPGMA rootless tree of the tested Chinese chestnut cultivars(Orange represents group 1,green represents group 2,and red represents an intermediate types between group 1 and group 2)
| 群体 Pop | 样品数 Sample | 位点数 Na | 有效等位位点 Ne | Shannon’s 多样性指数 I | 观察杂合度 Ho | 期望杂合度 He |
|---|---|---|---|---|---|---|
| Pop1(华北品种群North China group) | 190 | 8.714 ± 0.772 | 3.675 ± 0.282 | 1.450 ± 0.083 | 0.577 ± 0.032 | 0.685 ± 0.030 |
| Pop2(西北品种群Northwest China group) | 10 | 4.857 ± 0.347 | 3.286 ± 0.210 | 1.289 ± 0.080 | 0.595 ± 0.044 | 0.658 ± 0.032 |
| Pop3(长江中下游品种群Middle and lower reaches of the Yangtze River group) | 100 | 7.810 ± 0.699 | 3.320 ± 0.294 | 1.347 ± 0.094 | 0.601 ± 0.042 | 0.642 ± 0.035 |
| Pop4(东南品种群 Southeast China group) | 18 | 6.286 ± 0.522 | 3.593 ± 0.339 | 1.391 ± 0.096 | 0.582 ± 0.050 | 0.662 ± 0.035 |
| Pop5(西南品种群 Southwest China group) | 24 | 6.381 ± 0.567 | 3.368 ± 0.309 | 1.462 ± 0.091 | 0.585 ± 0.043 | 0.703 ± 0.029 |
表3 板栗5个品种群间的关键遗传数据
Table 3 The key genetic statistics of genetic diversity of five Chinese chestnut cultivar groups
| 群体 Pop | 样品数 Sample | 位点数 Na | 有效等位位点 Ne | Shannon’s 多样性指数 I | 观察杂合度 Ho | 期望杂合度 He |
|---|---|---|---|---|---|---|
| Pop1(华北品种群North China group) | 190 | 8.714 ± 0.772 | 3.675 ± 0.282 | 1.450 ± 0.083 | 0.577 ± 0.032 | 0.685 ± 0.030 |
| Pop2(西北品种群Northwest China group) | 10 | 4.857 ± 0.347 | 3.286 ± 0.210 | 1.289 ± 0.080 | 0.595 ± 0.044 | 0.658 ± 0.032 |
| Pop3(长江中下游品种群Middle and lower reaches of the Yangtze River group) | 100 | 7.810 ± 0.699 | 3.320 ± 0.294 | 1.347 ± 0.094 | 0.601 ± 0.042 | 0.642 ± 0.035 |
| Pop4(东南品种群 Southeast China group) | 18 | 6.286 ± 0.522 | 3.593 ± 0.339 | 1.391 ± 0.096 | 0.582 ± 0.050 | 0.662 ± 0.035 |
| Pop5(西南品种群 Southwest China group) | 24 | 6.381 ± 0.567 | 3.368 ± 0.309 | 1.462 ± 0.091 | 0.585 ± 0.043 | 0.703 ± 0.029 |

图4 板栗5个品种群的两两群体的Fst值 Pop1为华北品种群;Pop2为西北品种群;Pop3为长江中下游品种群;Pop4为东南品种群;Pop5为西南品种群。
Fig. 4 The Fst values of pairwise populations during five Chestnut Cultivars groups Pop1 is the north China group;Pop2 is the northwest China group;Pop3 is the middle and lower reaches of the Yangtze River group;Pop4 is the southeast China group;Pop5 is the southwest China group.
| 变异来源 Source of variance | 自由度 df | 方差总和 SS | 平均方差 MS | 变异组分 Variance component | 变异百分率/% Variation | P |
|---|---|---|---|---|---|---|
| 种群间Among Pops | 4 | 134.102 | 33.526 | 0.249 | 3 | < 0.001 |
| 个体间Among Indiv | 337 | 2722.196 | 8.078 | 0.965 | 13 | < 0.001 |
| 个体内Within Indiv | 342 | 2102.500 | 6.148 | 6.148 | 84 | < 0.010 |
| 总量Total | 683 | 4958.798 | 7.362 | 100 |
表4 5个板栗品种群的分子变异分析
Table 4 The AMOVA of five Chinese chestnut cultivar groups
| 变异来源 Source of variance | 自由度 df | 方差总和 SS | 平均方差 MS | 变异组分 Variance component | 变异百分率/% Variation | P |
|---|---|---|---|---|---|---|
| 种群间Among Pops | 4 | 134.102 | 33.526 | 0.249 | 3 | < 0.001 |
| 个体间Among Indiv | 337 | 2722.196 | 8.078 | 0.965 | 13 | < 0.001 |
| 个体内Within Indiv | 342 | 2102.500 | 6.148 | 6.148 | 84 | < 0.010 |
| 总量Total | 683 | 4958.798 | 7.362 | 100 |
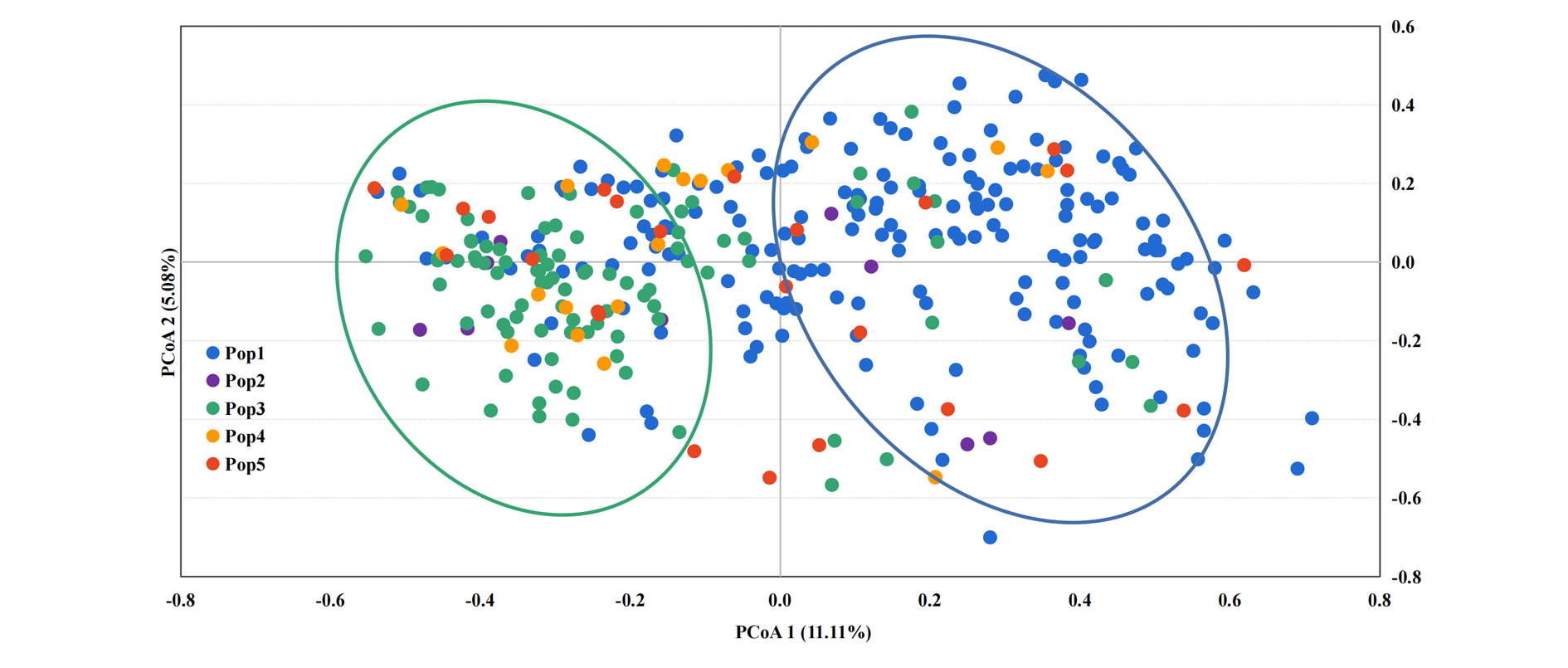
图5 板栗5个品种群的主坐标分析 Pop1为华北品种群;Pop2为西北品种群;Pop3为长江中下游品种群;Pop4为东南品种群;Pop5为西南品种群。
Fig. 5 Principal coordinate analysis of five Chinese chestnut cultivars groups Pop1 is the north China group;Pop2 is the northwest China group;Pop3 is the middle and lower reaches of the Yangtze River group;Pop4 is the southeast China group;Pop5 is the southwest China group.
| [1] |
Alcaraz M L, Hormaza J I. 2007. Molecular characterization and genetic diversity in an avocado collection of cultivars and local Spanish genotypes using SSRs. Hereditas, 144 (6):244-253.
doi: 10.1111/j.2007.0018-0661.02019x pmid: 18215247 |
| [2] | FAO. 2019. Value of agricultural production. http://faostat3.fao.org/. |
| [3] |
Falush D, Stephens M, Pritchard J K. 2003. Inference of population structure using multi-locus genotype data:Linked loci and correlated allele frequencies. Genetics, 164:1567-1587.
doi: 10.1093/genetics/164.4.1567 URL |
| [4] | Guo Jun, Zhu Jie, Xie Shangqian, Zhang Ye, Ye Beilei,Zheng Liyan,Ling Peng. 2020. Development of SSR molecular markers based on transcriptome and analysis of genetic relationship of germplasm resources in avocado. Acta Horticulturae Sinica, 47 (8):1552-1564. (in Chinese) |
| 郭俊, 朱婕, 谢尚潜, 张叶, 叶蓓蕾, 郑丽燕, 凌鹏. 2020. 油梨转录组SSR 分子标记开发与种质资源亲缘关系分析. 园艺学报, 47 (8):1552-1564. | |
| [5] | He Xu-dong, Zheng Ji-wei, Tian Xue-yao, Jiao Zhong-yi, Dou Quan-qin. 2021. Genetic relationship analysis and fingerprint construction of Carya illinoensis varieties. Forest Research, 34 (4):95-102. (in Chinese) |
| 何旭东, 郑纪伟, 田雪瑶, 教忠意, 窦全琴. 2021. 薄壳山核桃品种亲缘关系分析与指纹图谱构建. 林业科学研究, 34 (4):95-102. | |
| [6] |
Huang H W, Dane F, Norton J D. 1994. Allozyme diversity in Chinese Seguinii and American chestnut(Castanea spp.). Theoretical and Applied Genetics, 88 (8):981-985.
doi: 10.1007/BF00220805 pmid: 24186251 |
| [7] | Jiang Xibing, Tang Dan, Gong Bangchu, Lai Junsheng. 2015. Genetic diversity and association analysis of local cultivars of Chinese chestnut based on SSR markers. Acta Horticulturae Sinica, 42 (12):2478-2488. (in Chinese) |
| 江锡兵, 汤丹, 龚榜初, 赖俊声. 2015. 基于SSR标记的板栗地方品种遗传多样性与关联分析. 园艺学报, 42 (12):2478-2488. | |
| [8] | Jiang Xibing, Zhang Pingsheng, Xu Yang, Wu Conglian, Zhang Dongbei, Gong Bangchu, Wu Kaiyun, Lai Junsheng. 2021. Genetic diversity of F1 hybrids of chestnut based on SSR markers. Acta Horticulturae Sinica, 48 (5):897-907. (in Chinese) |
| 江锡兵, 章平生, 徐阳, 吴聪连, 张东北, 龚榜初, 吴开云, 赖俊声. 2021. 栗杂交F1代SSR标记遗传多样性分析. 园艺学报, 48 (5):897-907. | |
| [9] | Kubisiak T L, Nelson C D, Staton M E, Zhebentyayaeva T, Smith C, Olukolu B A, Fang C, Hebard F V, Anagnosakis S, Wheeler N, Sisco P H, Abbott A G, Sederoff R R. 2013. A transcriptome-based genetic map of Chinese chestnut(Castanea mollissima)and identification of regions of segmental homology with peach(Prunus persica). Tree Genetics & Genomes, 9:557-571. |
| [10] | Lang P, Dane F, Kubisiak T L. 2006. Phylogeny of Castanea(Fagaceae)based on chloroplasttrnT-L-F sequence data. Tree Genetics & Genomes, 2 (3):132-139. |
| [11] | Li Chao, Yang Ying, Chen Wei, Zheng Heyun, Liao Xinfu, Sun Yuping. 2022. Construction of DNA fingerprinting and clustering analysis with SSR markers for the muskmelon of Xizhoumi series. Acta Horticulturae Sinica, 49 (3):622-632. (in Chinese) |
| 李超, 杨英, 陈伟, 郑贺云, 廖新福, 孙玉萍. 2022. 西州密系列甜瓜SSR指纹图谱构建及聚类分析. 园艺学报, 49 (3):622-632. | |
| [12] | Liu Guo-bing, Cao Jun, Lan Yan-ping, Wang Jin-bao. 2017. Construction of SSR fingerprint on 33 ancient chestnut trees. Acta Agriculturae Universitatis Jiangxiensis, 39 (1):134-139. (in Chinese) |
| 刘国彬, 曹均, 兰彦平, 王金宝. 2017. 明清板栗古树指纹图谱的构建. 江西农业大学学报, 39 (1):134-139. | |
| [13] |
Liu K, Muse S V. 2005. PowerMarker:an integrated analysis environment for genetic marker analysis. Bioinformatics, 21:2128-2129.
doi: 10.1093/bioinformatics/bti282 URL |
| [14] |
Nie X H, Wang Z H, Liu N W, Song Li, Yan B Q, Xing Y, Zhang Q, Fang K F, Zhao Y L, Chen X, Wang G P, Qin L, Cao Q Q. 2021. Fingerprinting 146 Chinese chestnut accessions and selecting a core collection using SSR markers. Journal of Integrative Agriculture, 20 (5):1277-1286.
doi: 10.1016/S2095-3119(20)63400-1 |
| [15] | Nie Xing-hua, Zheng Rui-jie, Zhao Yong-lian, Cao Qing-qin, Qin Ling, Xing Yu. 2021. Genetic diversity evaluation of Castanea in China based on fluorescently labeled SSR. Scientia Agricultura Sinica, 54 (8):1739-1750. (in Chinese) |
| 聂兴华, 郑瑞杰, 赵永廉, 曹庆芹, 秦岭, 邢宇. 2021. 利用荧光SSR分子标记评估中国栗属植物遗传多样性. 中国农业科学, 54 (8):1739-1750. | |
| [16] |
Peakall R, Smouse P E. 2006. GENALEX 6:genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes, 6:288-295.
doi: 10.1111/j.1471-8286.2005.01155.x URL |
| [17] | Riva G A, Cyril J C, Yuval E, Leora S. 2000. Simple Sequence Repeats in Escherichia coli:abundance,distribution,composition,and polymorphism. Genome Research, 10 (1):62-71. |
| [18] | Rod P, Smouse P E. 2012. GenAlEx 6.5:genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics,(19):2537-2539. |
| [19] |
Selkoe K A, Toonen R J. 2006. Microsatellites for ecologists:a practical guide to using and evaluating microsatellite markers. Ecology Letters, 9:615-629.
doi: 10.1111/j.1461-0248.2006.00889.x URL |
| [20] |
Taberlet P, Luikart G. 1999. Non-invasive genetic sampling and individual identification. Biological Journal of the Linnean Society, 68:41-55.
doi: 10.1111/j.1095-8312.1999.tb01157.x URL |
| [21] |
Tian Hua, Kang Ming, Yao Xiao-Hong,Huang Hong-Wen. 2009. Genetic diversity in natural populations of Castanea mollissima inferred from nuclear SSR markers. Biodiversity Science, 17 (3):296-302. (in Chinese)
doi: 10.3724/SP.J.1003.2009.09043 URL |
|
田华, 康明, 李丽, 姚小洪, 黄宏文. 2009. 中国板栗自然居群微卫星(SSR)遗传多样性. 生物多样性, 17 (3):296-302.
doi: 10.3724/SP.J.1003.2009.09043 |
|
| [22] | UPOV. 2007. Guidelines for DNA-profiling:Molecular marker selection and database construction/BMT Guidelines(proj.9). Geneva,Switzerland:3-4. |
| [23] |
Waits L P, Luikart G, Taberlet P. 2001. Estimating the probability of identity among genotypes in natural populations:cautions and guidelines. Molecular Ecology, 10:249-256.
doi: 10.1046/j.1365-294x.2001.01185.x pmid: 11251803 |
| [24] |
Wright S. 1931. Evolution in Mendelian populations. Genetics, 16:97-159.
doi: 10.1093/genetics/16.2.97 pmid: 17246615 |
| [25] | Ye Shu-tao, Fang Zhou, Li Ying-lin, Wu Jun-jian, Zheng Guo-hua, Chen Hui, Li Yu. 2021. The SSR variation for cultivated population of Castanea henryi variety ‘Youzhen’. Molecular Plant Breeding, 6:1-9. (in Chinese) |
| 叶树涛, 方周, 李颖林, 吴钧剑, 郑国华, 陈辉, 李煜. 2021. 锥栗农家品种‘油榛’栽培群体的SSR变异分析. 分子植物育种, 6:1-9. | |
| [26] | Zhang He-yu, Liang Wei-jian. 2005. Chinese fruit tree,Chinese chestnut,hazelnut rolls. Beijing: China Forestry Publishing House. (in Chinese) |
| 张宇和, 柳鎏, 梁维坚. 2005. 中国果树志. 板栗,榛子卷. 北京: 中国林业出版社. | |
| [27] | Zhang Xin-fang, Zhang Shu-hang, Li Ying, Wang Guang-peng. 2020. Genetic diversity analysis of chestnut germplasm in Yanshan region based on SSR markers. Journal of China Agricultural University, 25 (4):61-71. (in Chinese) |
| 张馨方, 张树航, 李颖, 王广鹏. 2020. 基于SSR标记的燕山板栗种质资源遗传多样性分析. 中国农业大学学报, 25 (4):61-71. | |
| [28] | Zhou Lian-di. 2005. Study on genetic diversity of germplasm resources in Castanea mollissima[Ph. D. Dissertation]. China Agricultural University. (in Chinese) |
| 周连第. 2005. 板栗种质资源遗传多样性研究[博士论文]. 北京: 中国农业大学. |
| [1] | 王 瑞, 洪文娟, 罗 华, 赵丽娜, 陈 颖, 王 君, . 石榴品种SSR指纹图谱构建及杂种父本鉴定[J]. 园艺学报, 2023, 50(2): 265-278. |
| [2] | 任海龙, 许东林, 张 晶, 邹集文, 李光光, 周贤玉, 肖婉钰, 孙艺嘉. 菜薹KASP-SNP指纹图谱构建及品种鉴定[J]. 园艺学报, 2023, 50(2): 307-318. |
| [3] | 王梦梦, 孙德岭, 陈 锐, 杨迎霞, 张 冠, 吕明杰, 王 倩, 谢添羽, 牛国保, 单晓政, 谭 津, 姚星伟, . 花椰菜核心种质的构建与评价[J]. 园艺学报, 2023, 50(2): 421-431. |
| [4] | 刘艺平, 倪梦辉, 吴芳芳, 刘红利, 贺丹, 孔德政. 荷花花器官性状与SSR标记的关联分析[J]. 园艺学报, 2023, 50(1): 103-115. |
| [5] | 郭燕, 张树航, 李颖, 张馨方, 王英杰, 王广鹏. 燕山板栗种质资源叶片表型性状多样性研究[J]. 园艺学报, 2022, 49(8): 1673-1688. |
| [6] | 吕正鑫, 贺艳群, 贾东峰, 黄春辉, 钟敏, 廖光联, 朱壹, 袁开昌, 刘传浩, 徐小彪. 猕猴桃种质资源表型性状遗传多样性分析[J]. 园艺学报, 2022, 49(7): 1571-1581. |
| [7] | 李文婷, 李翠晓, 林小清, 郑永钦, 郑正, 邓晓玲. 基于STR位点对广东省柑橘溃疡病菌种群遗传结构的分析[J]. 园艺学报, 2022, 49(6): 1233-1246. |
| [8] | 张萌, 单玉莹, 杨业波, 翟飞飞, 王兆山, 巨关升, 孙振元, 李振坚. 中国石斛属植物遗传资源的AFLP分析[J]. 园艺学报, 2022, 49(6): 1339-1350. |
| [9] | 杨易, 黎庭耀, 李国景, 陈汉才, 沈卓, 周轩, 吴增祥, 吴新义, 张艳. 基于重测序的长豇豆基因组InDel标记开发及应用[J]. 园艺学报, 2022, 49(4): 778-790. |
| [10] | 李超, 杨英, 陈伟, 郑贺云, 廖新福, 孙玉萍. 西州密系列甜瓜SSR指纹图谱构建及聚类分析[J]. 园艺学报, 2022, 49(3): 622-632. |
| [11] | 李世帆, 邵宇纯, 祁豪男, 杨兰, 李小伟, 徐秉良, 陈雅寒. 甘肃省苹果坏死花叶病毒检测及遗传多样性分析[J]. 园艺学报, 2022, 49(11): 2431-2438. |
| [12] | 陈明堃, 陈璐, 孙维红, 马山虎, 兰思仁, 彭东辉, 刘仲健, 艾叶. 建兰种质资源遗传多样性分析及核心种质构建[J]. 园艺学报, 2022, 49(1): 175-186. |
| [13] | 宋芸, 贾孟君, 曹亚萍, 李政, 贺嘉欣, 王勇飞, 张鑫瑞, 乔永刚. 连翘叶绿体基因组特征分析[J]. 园艺学报, 2022, 49(1): 187-199. |
| [14] | 李艳红, 聂俊, 郑锦荣, 谭德龙, 张长远, 史亮亮, 谢玉明. 华南地区樱桃番茄表型性状遗传多样性分析及综合评价[J]. 园艺学报, 2021, 48(9): 1717-1730. |
| [15] | 王鑫, 李明阳, 田琳, 刘冬云. 利用ISSR标记及rDNA-ITS序列分析河北省野生铁线莲的亲缘关系[J]. 园艺学报, 2021, 48(9): 1755-1767. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司