
园艺学报 ›› 2022, Vol. 49 ›› Issue (8): 1637-1649.doi: 10.16420/j.issn.0513-353x.2021-0642
收稿日期:2022-01-07
修回日期:2022-05-30
出版日期:2022-08-25
发布日期:2022-09-05
通讯作者:
牛建新
E-mail:njx105@163.com
基金资助:
LIU Jinming, GUO Caihua, YUAN Xing, KANG Chao, QUAN Shaowen, NIU Jianxin*( )
)
Received:2022-01-07
Revised:2022-05-30
Online:2022-08-25
Published:2022-09-05
Contact:
NIU Jianxin
E-mail:njx105@163.com
摘要:
根据苹果、拟南芥等植物中的Dof转录因子基因序列,利用BLAST软件鉴定到梨基因组中51个PbDof基因,分为9个亚家族。基因结构分析表明,PbDof的内含子数为 1 ~ 3个;启动子区域含有5种胁迫响应和9种植物激素的顺式调控元件;基因复制结果显示,PbDof在物种进化过程中主要受纯化选择的作用。以蒸馏水为对照,于‘库尔勒香梨’盛花期喷施外源GA3和PP333,并采qRT-PCR技术检测各处理条件下宿萼组和脱萼组子房和萼片中PbDof的表达情况。结果表明,处理9 d后,PbDof34和PbDof51的表达量在GA3处理宿萼组萼片中均显著高于PP333处理脱萼组,且PbDof34显著高于对照;而PbDof16、PbDof19和PbDof44的表达量在GA3处理宿萼组萼片中均低于PP333处理脱萼组;此外,PbDof16和PbDof44的表达量在PP333处理脱萼组萼片中均显著高于对照。因此,PbDof34和PbDof51可能正向调控萼片宿存,PbDof16、PbDof19和PbDof44可能正向调控萼片脱落。
中图分类号:
刘金明, 郭彩华, 袁星, 亢超, 全绍文, 牛建新. 梨Dof家族基因鉴定及其在宿存与脱落萼片中的表达分析[J]. 园艺学报, 2022, 49(8): 1637-1649.
LIU Jinming, GUO Caihua, YUAN Xing, KANG Chao, QUAN Shaowen, NIU Jianxin. Genome-wide Identification of Dof Family Genes and Expression Analysis Sepal Persistent and Abscission in Pear[J]. Acta Horticulturae Sinica, 2022, 49(8): 1637-1649.
| 基因 | 上游引物(5′-3′) | 下游引物(5′-3′) | ||
|---|---|---|---|---|
| Gene | Forward primer | Reverse primer | ||
| PbDof11 | AACTCATGCCCTAAGCCTGC | TTGAACCACCTCCAACTGGG | ||
| PbDof16 | CCGGTGTTCTGTGGCCTTAT | CCCTCGAATGCTTCCCCAAT | ||
| PbDof19 | CTCCTTGGCCTTACCCTTGG | TCGAATGTTTCCCCAGGGTG | ||
| PbDof34 | AGGTCCAAAACGGCCAAGAT | CAGCCCTTGCAGAAGTGTCT | ||
| PbDof44 | CTCCTTGGCCTTACCCTTGG | CGGTGGAGCTTCTTCCTCAG | ||
| PbDof51 | CGGCACTTCTGCAAATCCTG | ACTCAACTGTAAGGCCGCTC | ||
| Actin | CCATCCAGGCTGTTCTCTC | GCAAGGTCCAGACGAAGG |
表1 荧光定量PCR引物
Table 1 Primer sequences for quantitative real time PCR
| 基因 | 上游引物(5′-3′) | 下游引物(5′-3′) | ||
|---|---|---|---|---|
| Gene | Forward primer | Reverse primer | ||
| PbDof11 | AACTCATGCCCTAAGCCTGC | TTGAACCACCTCCAACTGGG | ||
| PbDof16 | CCGGTGTTCTGTGGCCTTAT | CCCTCGAATGCTTCCCCAAT | ||
| PbDof19 | CTCCTTGGCCTTACCCTTGG | TCGAATGTTTCCCCAGGGTG | ||
| PbDof34 | AGGTCCAAAACGGCCAAGAT | CAGCCCTTGCAGAAGTGTCT | ||
| PbDof44 | CTCCTTGGCCTTACCCTTGG | CGGTGGAGCTTCTTCCTCAG | ||
| PbDof51 | CGGCACTTCTGCAAATCCTG | ACTCAACTGTAAGGCCGCTC | ||
| Actin | CCATCCAGGCTGTTCTCTC | GCAAGGTCCAGACGAAGG |
| 基因 Gene | ID | 染色体位置 Chromosome location | 染色体重命名 Chromosome rename | 长度/aa Length | 分子量/kD MW | 等电点 pI | α-螺旋/% α-helix | β-转角/% β-turn | 无规则卷曲/% Random coil | 延长链/% Extended strand |
|---|---|---|---|---|---|---|---|---|---|---|
| PbDof1 | XM_009365818.2 | Scaffold3.0 | Scaff1 | 248 | 26.31 | 5.95 | 8.87 | 4.44 | 70.97 | 15.73 |
| PbDof2 | XM_009368130.2 | Scaffold4.0 | Scaff2 | 387 | 41.62 | 9.33 | 17.83 | 3.88 | 66.41 | 11.89 |
| PbDof3 | XM_018649999.1 | Scaffold4.0 | Scaff2 | 239 | 24.85 | 8.64 | 8.37 | 7.11 | 66.95 | 17.57 |
| PbDof4 | XM_009354127.2 | Scaffold21.0 | Scaff3 | 304 | 33.58 | 6.45 | 15.13 | 3.62 | 64.47 | 16.78 |
| PbDof5 | XM_009354128.2 | Scaffold21.0 | Scaff3 | 304 | 33.58 | 6.45 | 15.13 | 3.62 | 64.47 | 16.78 |
| PbDof6 | XM_009354499.2 | Scaffold24.0 | Scaff4 | 163 | 18.27 | 9.43 | 10.43 | 4.91 | 68.71 | 15.95 |
| PbDof7 | XM_009354615.2 | Scaffold24.0 | Scaff4 | 378 | 41.07 | 8.05 | 13.23 | 0.79 | 77.78 | 8.20 |
| PbDof8 | XM_009354693.2 | Scaffold24.0 | Scaff4 | 290 | 31.20 | 8.51 | 11.72 | 3.45 | 75.17 | 9.66 |
| PbDof9 | XM_009355416.2 | Scaffold31.0 | Scaff5 | 368 | 39.79 | 8.59 | 8.97 | 3.80 | 75.82 | 11.41 |
| PbDof10 | XM_009359983.2 | Scaffold63.0 | Scaff6 | 291 | 32.10 | 7.64 | 15.81 | 3.09 | 67.35 | 13.75 |
| PbDof11 | XM_009360389.2 | Scaffold67.0 | Scaff7 | 314 | 34.38 | 9.33 | 9.24 | 2.23 | 77.71 | 10.83 |
| 基因 Gene | ID | 染色体位置 Chromosome location | 染色体重命名 Chromosome rename | 长度/aa Length | 分子量/kD MW | 等电点 pI | α-螺旋/% α-helix | β-转角/% β-turn | 无规则卷曲/% Random coil | 延长链/% Extended strand |
| PbDof12 | XM_009361313.1 | Scaffold75.0 | Scaff8 | 327 | 36.06 | 9.00 | 7.95 | 1.53 | 74.62 | 15.90 |
| PbDof13 | XM_018648309.1 | Scaffold83.0 | Scaff9 | 523 | 55.98 | 5.94 | 13.77 | 2.29 | 76.67 | 7.27 |
| PbDof14 | XM_018648376.1 | Scaffold84.0 | Scaff10 | 302 | 33.89 | 5.11 | 25.50 | 1.66 | 66.89 | 5.96 |
| PbDof15 | XM_009364743.2 | Scaffold106.0 | Scaff11 | 327 | 36.23 | 6.69 | 12.54 | 2.75 | 66.36 | 18.35 |
| PbDof16 | XM_009371010.2 | Scaffold180.0 | Scaff12 | 465 | 50.96 | 5.79 | 12.04 | 1.94 | 79.78 | 6.24 |
| PbDof17 | XM_009377147.2 | Scaffold275.0 | Scaff13 | 286 | 31.52 | 7.71 | 5.59 | 1.05 | 80.42 | 12.94 |
| PbDof18 | XM_009377148.2 | Scaffold275.0 | Scaff13 | 270 | 29.76 | 8.44 | 6.30 | 3.33 | 77.78 | 12.59 |
| PbDof19 | XM_009378003.2 | Scaffold292.0 | Scaff14 | 511 | 55.22 | 5.72 | 10.37 | 1.37 | 80.82 | 7.44 |
| PbDof20 | XM_009336089.2 | Scaffold384.0 | Scaff15 | 289 | 31.75 | 7.68 | 8.30 | 1.73 | 80.28 | 9.69 |
| PbDof21 | XM_009340481.2 | Scaffold499.0 | Scaff16 | 366 | 38.92 | 9.13 | 9.29 | 3.83 | 77.60 | 9.29 |
| PbDof22 | XM_009344990.1 | Scaffold698.0 | Scaff17 | 316 | 35.40 | 6.43 | 12.03 | 2.53 | 74.68 | 10.76 |
| PbDof23 | XM_009344991.1 | Scaffold698.0 | Scaff17 | 316 | 34.97 | 6.52 | 13.61 | 2.85 | 72.78 | 10.76 |
| PbDof24 | XM_009345555.2 | Scaffold727.0 | Scaff18 | 372 | 40.05 | 8.78 | 8.06 | 2.42 | 79.84 | 9.68 |
| PbDof25 | XM_009345556.2 | Scaffold727.0 | Scaff18 | 337 | 36.26 | 8.96 | 6.53 | 2.97 | 81.31 | 9.20 |
| PbDof26 | XM_009346163.2 | Scaffold765.0 | Scaff19 | 225 | 24.05 | 8.18 | 6.67 | 3.56 | 79.11 | 10.67 |
| PbDof27 | XM_009346170.2 | Scaffold765.0 | Scaff19 | 225 | 24.05 | 8.18 | 6.67 | 3.56 | 79.11 | 10.67 |
| PbDof28 | XM_009348304.2 | Scaffold915.0 | Scaff20 | 304 | 33.60 | 6.62 | 18.42 | 1.64 | 64.80 | 15.13 |
| PbDof29 | XM_009348967.2 | Scaffold973.0 | Scaff21 | 347 | 37.14 | 8.90 | 20.46 | 2.59 | 66.28 | 10.66 |
| PbDof30 | XM_009349812.2 | Scaffold1047.0 | Scaff22 | 346 | 36.91 | 8.84 | 19.65 | 3.18 | 65.32 | 11.85 |
| PbDof31 | XM_009352168.2 | Scaffold1355.0 | Scaff23 | 303 | 33.81 | 4.94 | 20.79 | 1.65 | 69.64 | 7.92 |
| PbDof32 | XM_009336822.2 | Scaffold1.0.1 | Scaff24 | 291 | 32.49 | 7.70 | 15.81 | 2.06 | 74.23 | 7.90 |
| PbDof33 | XM_009343730.2 | Scaffold1.0.1 | Scaff24 | 378 | 41.13 | 8.58 | 15.08 | 0.53 | 76.19 | 8.20 |
| PbDof34 | XM_009351106.2 | Scaffold1.0.1 | Scaff24 | 163 | 18.34 | 9.08 | 14.11 | 6.13 | 63.19 | 16.56 |
| PbDof35 | XM_009354801.2 | Scaffold1.0.1 | Scaff24 | 291 | 31.31 | 9.30 | 16.84 | 5.50 | 63.23 | 14.43 |
| PbDof36 | XM_009339603.2 | Scaffold14.0.1 | Scaff25 | 323 | 34.90 | 7.20 | 15.17 | 2.79 | 69.04 | 13.00 |
| PbDof37 | XM_009340881.2 | Scaffold14.0.1 | Scaff25 | 463 | 50.48 | 5.48 | 13.17 | 1.51 | 76.89 | 8.42 |
| PbDof38 | XM_009345020.2 | Scaffold16.0.1 | Scaff26 | 362 | 39.20 | 8.99 | 9.12 | 4.14 | 73.76 | 12.98 |
| PbDof39 | XM_009347952.2 | Scaffold30.0.1 | Scaff27 | 325 | 34.81 | 7.21 | 8.31 | 1.85 | 76.00 | 13.85 |
| PbDof40 | XM_009351662.2 | Scaffold17.0.2 | Scaff28 | 282 | 31.49 | 6.77 | 10.64 | 3.90 | 74.11 | 11.35 |
| PbDof41 | XM_009353689.2 | Scaffold20.0.1 | Scaff29 | 320 | 35.00 | 7.23 | 10.94 | 2.50 | 75.94 | 10.62 |
| PbDof42 | XM_009359445.2 | Scaffold58.0.1 | Scaff30 | 357 | 38.71 | 8.93 | 10.36 | 4.76 | 71.43 | 13.45 |
| PbDof43 | XM_009362338.2 | Scaffold80.0.1 | Scaff31 | 539 | 58.63 | 6.66 | 13.17 | 1.67 | 78.11 | 7.05 |
| PbDof44 | XM_009362339.2 | Scaffold80.0.1 | Scaff31 | 521 | 56.57 | 6.22 | 13.05 | 2.30 | 77.93 | 6.72 |
| PbDof45 | XM_009363055.2 | Scaffold87.0.1 | Scaff32 | 316 | 34.47 | 8.16 | 9.18 | 4.43 | 74.05 | 12.34 |
| PbDof46 | XM_009365959.2 | Scaffold116.0.1 | Scaff33 | 428 | 45.70 | 9.03 | 16.12 | 2.57 | 72.43 | 8.88 |
| PbDof47 | XM_009366523.2 | Scaffold122.0.1 | Scaff34 | 384 | 40.83 | 8.75 | 15.10 | 2.08 | 73.96 | 8.85 |
| PbDof48 | XM_009367372.2 | Scaffold131.0.1 | Scaff35 | 253 | 26.84 | 6.15 | 14.62 | 7.51 | 55.34 | 22.53 |
| PbDof49 | XM_009369256.2 | Scaffold155.0.1 | Scaff36 | 221 | 23.59 | 8.13 | 10.86 | 2.71 | 76.02 | 10.41 |
| PbDof50 | XM_009376412.2 | Scaffold262.0.1 | Scaff37 | 376 | 40.32 | 9.64 | 13.83 | 3.99 | 74.20 | 7.98 |
| PbDof51 | XM_009378209.2 | Scaffold294.0.1 | Scaff38 | 247 | 25.39 | 8.18 | 9.31 | 5.67 | 67.61 | 17.41 |
表2 梨的Dof基因家族
Table 2 The Dof gene family of pear
| 基因 Gene | ID | 染色体位置 Chromosome location | 染色体重命名 Chromosome rename | 长度/aa Length | 分子量/kD MW | 等电点 pI | α-螺旋/% α-helix | β-转角/% β-turn | 无规则卷曲/% Random coil | 延长链/% Extended strand |
|---|---|---|---|---|---|---|---|---|---|---|
| PbDof1 | XM_009365818.2 | Scaffold3.0 | Scaff1 | 248 | 26.31 | 5.95 | 8.87 | 4.44 | 70.97 | 15.73 |
| PbDof2 | XM_009368130.2 | Scaffold4.0 | Scaff2 | 387 | 41.62 | 9.33 | 17.83 | 3.88 | 66.41 | 11.89 |
| PbDof3 | XM_018649999.1 | Scaffold4.0 | Scaff2 | 239 | 24.85 | 8.64 | 8.37 | 7.11 | 66.95 | 17.57 |
| PbDof4 | XM_009354127.2 | Scaffold21.0 | Scaff3 | 304 | 33.58 | 6.45 | 15.13 | 3.62 | 64.47 | 16.78 |
| PbDof5 | XM_009354128.2 | Scaffold21.0 | Scaff3 | 304 | 33.58 | 6.45 | 15.13 | 3.62 | 64.47 | 16.78 |
| PbDof6 | XM_009354499.2 | Scaffold24.0 | Scaff4 | 163 | 18.27 | 9.43 | 10.43 | 4.91 | 68.71 | 15.95 |
| PbDof7 | XM_009354615.2 | Scaffold24.0 | Scaff4 | 378 | 41.07 | 8.05 | 13.23 | 0.79 | 77.78 | 8.20 |
| PbDof8 | XM_009354693.2 | Scaffold24.0 | Scaff4 | 290 | 31.20 | 8.51 | 11.72 | 3.45 | 75.17 | 9.66 |
| PbDof9 | XM_009355416.2 | Scaffold31.0 | Scaff5 | 368 | 39.79 | 8.59 | 8.97 | 3.80 | 75.82 | 11.41 |
| PbDof10 | XM_009359983.2 | Scaffold63.0 | Scaff6 | 291 | 32.10 | 7.64 | 15.81 | 3.09 | 67.35 | 13.75 |
| PbDof11 | XM_009360389.2 | Scaffold67.0 | Scaff7 | 314 | 34.38 | 9.33 | 9.24 | 2.23 | 77.71 | 10.83 |
| 基因 Gene | ID | 染色体位置 Chromosome location | 染色体重命名 Chromosome rename | 长度/aa Length | 分子量/kD MW | 等电点 pI | α-螺旋/% α-helix | β-转角/% β-turn | 无规则卷曲/% Random coil | 延长链/% Extended strand |
| PbDof12 | XM_009361313.1 | Scaffold75.0 | Scaff8 | 327 | 36.06 | 9.00 | 7.95 | 1.53 | 74.62 | 15.90 |
| PbDof13 | XM_018648309.1 | Scaffold83.0 | Scaff9 | 523 | 55.98 | 5.94 | 13.77 | 2.29 | 76.67 | 7.27 |
| PbDof14 | XM_018648376.1 | Scaffold84.0 | Scaff10 | 302 | 33.89 | 5.11 | 25.50 | 1.66 | 66.89 | 5.96 |
| PbDof15 | XM_009364743.2 | Scaffold106.0 | Scaff11 | 327 | 36.23 | 6.69 | 12.54 | 2.75 | 66.36 | 18.35 |
| PbDof16 | XM_009371010.2 | Scaffold180.0 | Scaff12 | 465 | 50.96 | 5.79 | 12.04 | 1.94 | 79.78 | 6.24 |
| PbDof17 | XM_009377147.2 | Scaffold275.0 | Scaff13 | 286 | 31.52 | 7.71 | 5.59 | 1.05 | 80.42 | 12.94 |
| PbDof18 | XM_009377148.2 | Scaffold275.0 | Scaff13 | 270 | 29.76 | 8.44 | 6.30 | 3.33 | 77.78 | 12.59 |
| PbDof19 | XM_009378003.2 | Scaffold292.0 | Scaff14 | 511 | 55.22 | 5.72 | 10.37 | 1.37 | 80.82 | 7.44 |
| PbDof20 | XM_009336089.2 | Scaffold384.0 | Scaff15 | 289 | 31.75 | 7.68 | 8.30 | 1.73 | 80.28 | 9.69 |
| PbDof21 | XM_009340481.2 | Scaffold499.0 | Scaff16 | 366 | 38.92 | 9.13 | 9.29 | 3.83 | 77.60 | 9.29 |
| PbDof22 | XM_009344990.1 | Scaffold698.0 | Scaff17 | 316 | 35.40 | 6.43 | 12.03 | 2.53 | 74.68 | 10.76 |
| PbDof23 | XM_009344991.1 | Scaffold698.0 | Scaff17 | 316 | 34.97 | 6.52 | 13.61 | 2.85 | 72.78 | 10.76 |
| PbDof24 | XM_009345555.2 | Scaffold727.0 | Scaff18 | 372 | 40.05 | 8.78 | 8.06 | 2.42 | 79.84 | 9.68 |
| PbDof25 | XM_009345556.2 | Scaffold727.0 | Scaff18 | 337 | 36.26 | 8.96 | 6.53 | 2.97 | 81.31 | 9.20 |
| PbDof26 | XM_009346163.2 | Scaffold765.0 | Scaff19 | 225 | 24.05 | 8.18 | 6.67 | 3.56 | 79.11 | 10.67 |
| PbDof27 | XM_009346170.2 | Scaffold765.0 | Scaff19 | 225 | 24.05 | 8.18 | 6.67 | 3.56 | 79.11 | 10.67 |
| PbDof28 | XM_009348304.2 | Scaffold915.0 | Scaff20 | 304 | 33.60 | 6.62 | 18.42 | 1.64 | 64.80 | 15.13 |
| PbDof29 | XM_009348967.2 | Scaffold973.0 | Scaff21 | 347 | 37.14 | 8.90 | 20.46 | 2.59 | 66.28 | 10.66 |
| PbDof30 | XM_009349812.2 | Scaffold1047.0 | Scaff22 | 346 | 36.91 | 8.84 | 19.65 | 3.18 | 65.32 | 11.85 |
| PbDof31 | XM_009352168.2 | Scaffold1355.0 | Scaff23 | 303 | 33.81 | 4.94 | 20.79 | 1.65 | 69.64 | 7.92 |
| PbDof32 | XM_009336822.2 | Scaffold1.0.1 | Scaff24 | 291 | 32.49 | 7.70 | 15.81 | 2.06 | 74.23 | 7.90 |
| PbDof33 | XM_009343730.2 | Scaffold1.0.1 | Scaff24 | 378 | 41.13 | 8.58 | 15.08 | 0.53 | 76.19 | 8.20 |
| PbDof34 | XM_009351106.2 | Scaffold1.0.1 | Scaff24 | 163 | 18.34 | 9.08 | 14.11 | 6.13 | 63.19 | 16.56 |
| PbDof35 | XM_009354801.2 | Scaffold1.0.1 | Scaff24 | 291 | 31.31 | 9.30 | 16.84 | 5.50 | 63.23 | 14.43 |
| PbDof36 | XM_009339603.2 | Scaffold14.0.1 | Scaff25 | 323 | 34.90 | 7.20 | 15.17 | 2.79 | 69.04 | 13.00 |
| PbDof37 | XM_009340881.2 | Scaffold14.0.1 | Scaff25 | 463 | 50.48 | 5.48 | 13.17 | 1.51 | 76.89 | 8.42 |
| PbDof38 | XM_009345020.2 | Scaffold16.0.1 | Scaff26 | 362 | 39.20 | 8.99 | 9.12 | 4.14 | 73.76 | 12.98 |
| PbDof39 | XM_009347952.2 | Scaffold30.0.1 | Scaff27 | 325 | 34.81 | 7.21 | 8.31 | 1.85 | 76.00 | 13.85 |
| PbDof40 | XM_009351662.2 | Scaffold17.0.2 | Scaff28 | 282 | 31.49 | 6.77 | 10.64 | 3.90 | 74.11 | 11.35 |
| PbDof41 | XM_009353689.2 | Scaffold20.0.1 | Scaff29 | 320 | 35.00 | 7.23 | 10.94 | 2.50 | 75.94 | 10.62 |
| PbDof42 | XM_009359445.2 | Scaffold58.0.1 | Scaff30 | 357 | 38.71 | 8.93 | 10.36 | 4.76 | 71.43 | 13.45 |
| PbDof43 | XM_009362338.2 | Scaffold80.0.1 | Scaff31 | 539 | 58.63 | 6.66 | 13.17 | 1.67 | 78.11 | 7.05 |
| PbDof44 | XM_009362339.2 | Scaffold80.0.1 | Scaff31 | 521 | 56.57 | 6.22 | 13.05 | 2.30 | 77.93 | 6.72 |
| PbDof45 | XM_009363055.2 | Scaffold87.0.1 | Scaff32 | 316 | 34.47 | 8.16 | 9.18 | 4.43 | 74.05 | 12.34 |
| PbDof46 | XM_009365959.2 | Scaffold116.0.1 | Scaff33 | 428 | 45.70 | 9.03 | 16.12 | 2.57 | 72.43 | 8.88 |
| PbDof47 | XM_009366523.2 | Scaffold122.0.1 | Scaff34 | 384 | 40.83 | 8.75 | 15.10 | 2.08 | 73.96 | 8.85 |
| PbDof48 | XM_009367372.2 | Scaffold131.0.1 | Scaff35 | 253 | 26.84 | 6.15 | 14.62 | 7.51 | 55.34 | 22.53 |
| PbDof49 | XM_009369256.2 | Scaffold155.0.1 | Scaff36 | 221 | 23.59 | 8.13 | 10.86 | 2.71 | 76.02 | 10.41 |
| PbDof50 | XM_009376412.2 | Scaffold262.0.1 | Scaff37 | 376 | 40.32 | 9.64 | 13.83 | 3.99 | 74.20 | 7.98 |
| PbDof51 | XM_009378209.2 | Scaffold294.0.1 | Scaff38 | 247 | 25.39 | 8.18 | 9.31 | 5.67 | 67.61 | 17.41 |
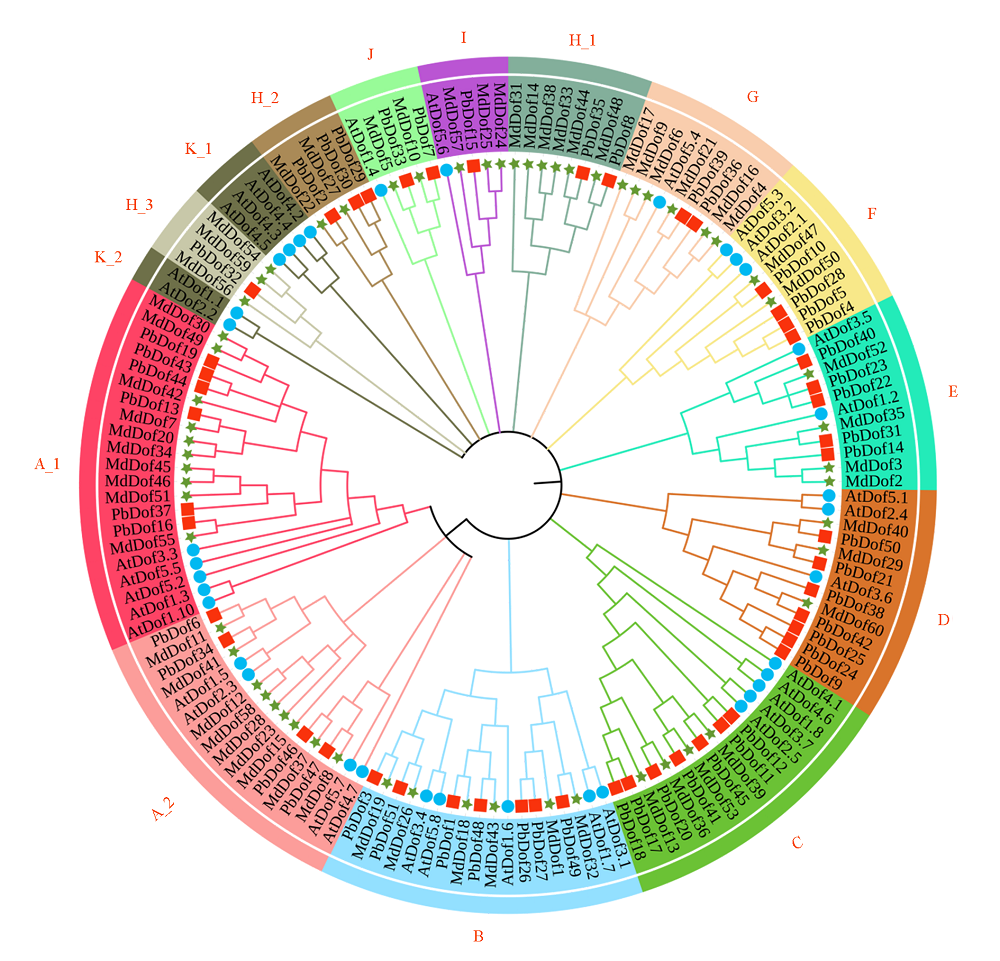
图1 拟南芥、苹果和梨Dof基因家族的系统发育关系 Pb:梨;Md:苹果;At:拟南芥。
Fig. 1 The phylogenetic relationship of the Dof gene family of Arabidopsis,apple and pear Pb:Pyrus × bretschneideri;Md:Malus domestica;At:Arabidopsis thaliana.
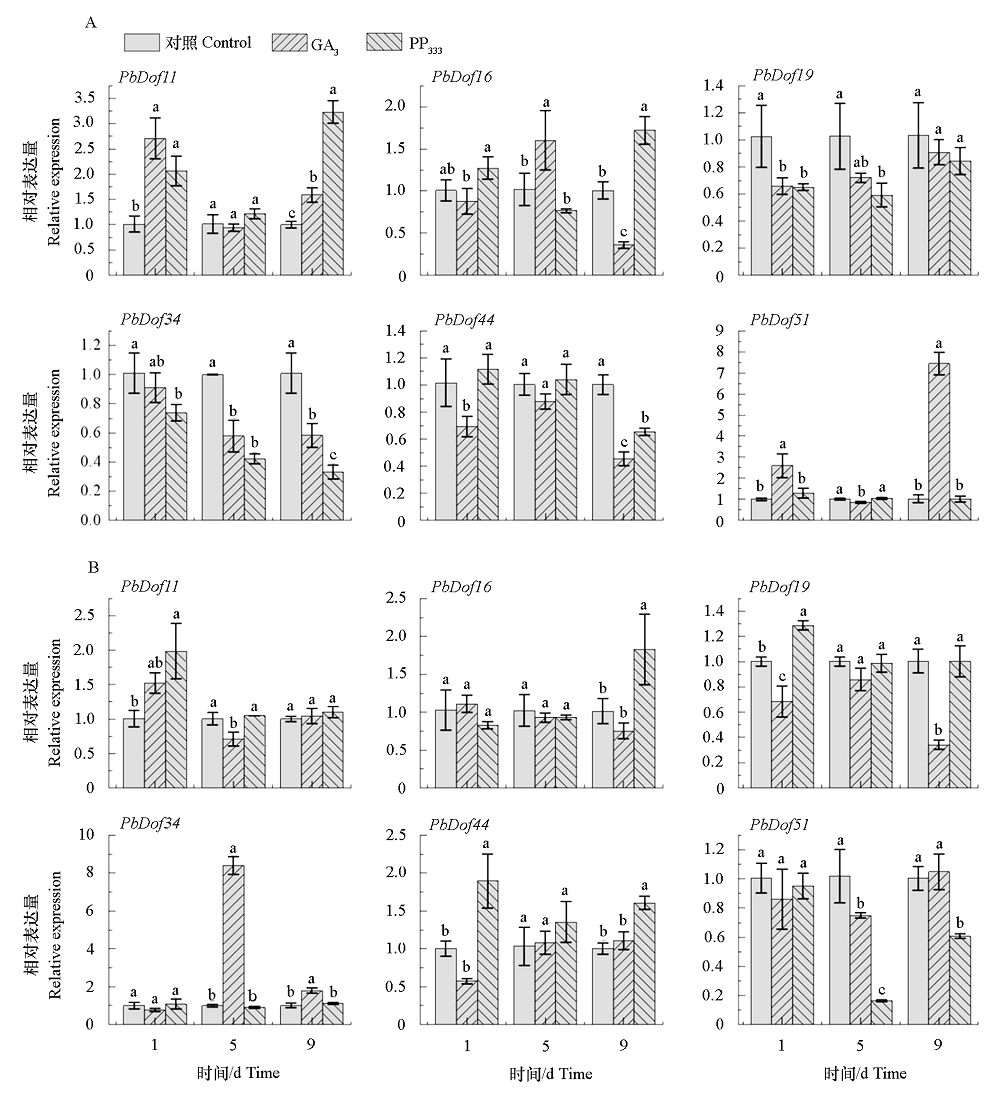
图5 PbDof在子房(A)和萼片(B)的相对表达量 采用邓肯氏多重比较;同一处理时间不同小写字母表示处理间差异显著(P < 0.05)。
Fig. 5 The relative expression levels of the PbDofs in ovary(A)and sepal(B) Multiple comparison using Duncan’s;different lowercase letters at the same time indicate significant differences at the level of P < 0.05 between treatments.
| [1] | Cai M, Lin J, Li Z, Lin Z, Ming R. 2020. Allele specific expression of Dof genes responding to hormones and abiotic stresses in sugarcane. PLoS ONE, 15 (1):e0227716. |
| [2] |
Cai X, Zhang Y, Zhang C, Zhang T, Hu T, Ye J, Zhang J, Wang T, Li H, Ye Z. 2013. Genome-wide analysis of plant-specific Dof transcription factor family in tomato. Journal of Integrative Plant Biology, 55 (6):552-566.
doi: 10.1111/jipb.12043 URL |
| [3] |
Chen P, Yan M, Li L, He J, Zhou S, Li Z, Niu C, Bao C, Zhi F, Ma F, Guan Q. 2020. The apple DNA-binding one zinc-finger protein MdDof 54 promotes drought resistance. Hortic Res, 7 (1):195.
doi: 10.1038/s41438-020-00419-5 URL |
| [4] |
Gupta S, Malviya N, Kushwaha H, Nasim J, Bisht N C, Singh V K, Yadav D. 2015. Insights into structural and functional diversity of Dof(DNA binding with one finger)transcription factor. Planta, 241 (3):549-562.
doi: 10.1007/s00425-014-2239-3 pmid: 25564353 |
| [5] | He Zishun, Niu Jianxin, Wu Zhonghua, Qin Weiming, Zhao Jianshe. 2007. A study on development law of calyx of Korla Fragrant Pear(Pyrus brestschneideri Rehd). Xinjiang Agricultural Sciences, 44 (3):377-381. (in Chinese) |
| 何子顺, 牛建新, 吴忠华, 覃伟铭, 赵建设. 2007. 库尔勒香梨花萼发育规律研究. 新疆农业科学, 44 (3):377-381. | |
| [6] | Iwamoto M, Higo K, Takano M. 2009. Circadian clock- and phytochrome-regulated Dof-like gene,Rdd1,is associated with grain size in rice. Plant,Cell & Environment, 32 (1):592-603. |
| [7] | Jia Bing, Guo Guoling, Wang Youyu, Wei Pengfei, Yu Tao, Chang Xiao, Heng Wei. 2021. Relationship between morphogenesis and carbohydrate synthesis of the calyx-persistence fruit and calyx-shedding fruit in differentiation stage of‘Dangshan Suli’pear. Acta Horticulturae Sinica, 48 (3):421-438. (in Chinese) |
| 贾兵, 郭国凌, 王友煜, 魏鹏飞, 余桃, 常笑, 衡伟. 2021. ‘砀山酥梨’宿萼和脱萼分化期形态建成与碳水化合物的关系. 园艺学报, 48 (3):421-438. | |
| [8] | Jiang Yanchen. 2011. Research of differences of endogenous hormones and quality in calyx persistent or fall off fruits[Ph. D. Dissertation]. Nanjing: Nanjing Agricultural University. (in Chinese) |
| 姜彦辰. 2011. 梨萼片脱落与宿存果实内源激素及品质的差异研究[博士论文]. 南京: 南京农业大学. | |
| [9] |
Kushwaha H, Gupta S, Singh V K, Rastogi S, Yadav D. 2011. Genome wide identification of Dof transcription factor gene family in sorghum and its comparative phylogenetic analysis with rice and Arabidopsis. Molecular Biology Reports, 38 (8):5037-5053.
doi: 10.1007/s11033-010-0650-9 pmid: 21161392 |
| [10] |
Lee Y, Yoon T H, Lee J, Jeon S Y, Lee J H, Lee M K, Chen H, Yun J, Oh S Y, Wen X, Cho H K, Mang H, Kwak J M. 2018. A lignin molecular brace controls precision processing of cell walls critical for surface integrity in Arabidopsis. Cell, 173 (6):1468-1480.
doi: 10.1016/j.cell.2018.03.060 |
| [11] | Li Changjiang, Li Peng, Jing Chunzhi, Tian Jia, Zhang Yuan, Li Jiang. 2017. The relationship between endogenous hormones distribution in fruitlets and calyx shedding of‘Korla Fragrant Pear’. Acta Agriculturae Boreali-Occidentalis Sinica, 26 (11):1631-1638. (in Chinese) |
| 李长江, 李鹏, 井春芝, 田嘉, 张渊, 李疆. 2017. ‘库尔勒香梨’幼果内源激素分布差异与果实萼片脱落的关系. 西北农业学报, 26 (11):1631-1638. | |
| [12] |
Lijavetzky D, Carbonero P, Vicente-Carbajosa J. 2003. Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evolutionary Biology, 3 (1):17.
doi: 10.1186/1471-2148-3-17 URL |
| [13] |
Liu J, Cheng Z, Xie L, Li X, Gao J. 2019. Multifaceted role of PheDof12- 1 in the regulation of flowering time and abiotic stress responses in moso bamboo(Phyllostachys edulis). International Journal of Molecular Sciences, 20 (2):424. doi: 10.3390/ijms20020424.
doi: 10.3390/ijms20020424 |
| [14] | Liu Ni, Tao Shutian, Li Leiyan, Huang Wenjiang, Zhang Shaoling. 2013. Changes in endogenous hormones levels of young fruit of‘Dangshan Suli’(Pyrus bretschneideri Rehd.)pear during calyx abscission processes. Journal of Nanjing Agricultural University, 36 (6):147-150. (in Chinese) |
| 刘妮, 陶书田, 李雷廷, 黄文江, 张绍玲. 2013. ‘砀山酥梨’幼果萼片脱落期内源激素含量变化. 南京农业大学学报, 36 (6):147-150. | |
| [15] |
Ma J, Li M Y, Wang F, Tang J, Xiong A S. 2015. Genome-wide analysis of Dof family transcription factors and their responses to abiotic stresses in Chinese cabbage. BMC Genomics, 16 (1):33.
doi: 10.1186/s12864-015-1242-9 URL |
| [16] | Ma Li, Zhou Li, Xu Hang, Quan Shaowen, Yang Jieping, Niu Jianxin. 2019. Evolutionary characteristics and the expression patterns of miR159 gene family in‘Kuerlexiangli’pear. Journal of Fruit Science, 36 (1):1-10. (in Chinese) |
| 马丽, 周丽, 徐航, 全绍文, 杨洁萍, 牛建新. 2019. ‘库尔勒香梨’miR159家族成员进化特性及表达分析. 果树学报, 36 (1):1-10. | |
| [17] |
Moreno-Risueno M A, Día I, Carrillo L, Fuentes R, Carbonero P. 2007. The HvDof 19 transcription factor mediates the abscisic acid-dependent repression of hydrolase genes in germinating barley aleurone. The Plant Journal:for Cell and Molecular Biology, 51 (3):352-365.
doi: 10.1111/j.1365-313X.2007.03146.x URL |
| [18] | Noguero M, Atif R M, Ochatt S, Thompson R D. 2013. The role of the DNA-binding one zinc finger(Dof)transcription factor family in plants. Plant Science:an International Journal of Experimental Plant Biology, 209:32-45. |
| [19] |
Pei M, Niu J, Li C, Cao F, Quan S. 2016. Identification and expression analysis of genes related to calyx persistence in Korla fragrant pear. BMC Genomics, 17:132.
doi: 10.1186/s12864-016-2470-3 URL |
| [20] | Qi Xiaoxiao. 2014. Investigation of genes expression of calyx survival and shedding of pear by digital gene expression and functional analysis of PsIDA and PsJOINTLESS[Ph. D. Dissertation]. Nanjing: Nanjing Agricultural University. (in Chinese) |
| 齐笑笑. 2014. 梨果实萼片宿存与脱落过程基因表达谱分析及PsIDA、PsJOINTLESS基因功能的初步研究[博士论文]. 南京: 南京农业大学. | |
| [21] | Qiao Yonggang, Wang Yongfei, Cao Yaping, He Jiaxin, Jia Mengjun, Li Zheng, Zhang Xinrui, Song Yun. 2020. Reference genes selection and related genes expression analysis under low and high temperature stress in Taraxacum officinale. Acta Horticulturae Sinica, 47 (6):1153-1164. (in Chinese) |
| 乔永刚, 王勇飞, 曹亚萍, 贺嘉欣, 贾孟君, 李政, 张鑫瑞, 宋芸. 2020. 药用蒲公英低温和高温胁迫下内参基因筛选与相关基因表达分析. 园艺学报, 47 (6):1153-1164. | |
| [22] |
Rojas-Gracia P, Roque E, Medina M, López-Martín M J, Cañas L A, Beltrán J P, Gómez-Mena C. 2019. The Dof transcription factor SlDof 10 regulates vascular tissue formation during ovary development in tomato. Frontiers in Plant Science, 10:216. doi: 10.3389/fpls.2019.00216.
doi: 10.3389/fpls.2019.00216 |
| [23] | Su J, Jia B, Jia S, Ye Z F, Heng W, Zhu L W. 2015. Effect of plant growth regulators on calyx abscission,fruit quality,and auxin-repressed protein (ARP)gene expression in fruitlets of‘Dangshan Suli’pear(Pyrus bretschneideri Rehd.). Journal of Horticultural Science & Biotechnology, 90 (2):135-142. |
| [24] |
Umemura Y, Ishiduka T, Yamamoto R, Esaka M. 2004. The Dof domain, a zinc finger DNA-binding domain conserved only in higher plants,truly functions as a Cys2/Cys 2 Zn finger domain. The Plant Journal:for Cell and Molecular Biology, 37 (5):741-749.
doi: 10.1111/j.1365-313X.2003.01997.x URL |
| [25] |
Washio K. 2001. Identification of Dof proteins with implication in the gibberellin-regulated expression of a peptidase gene following the germination of rice grains. Biochimica et Biophysica Acta, 1520 (1):54-62.
pmid: 11470159 |
| [26] |
Washio K. 2003. Functional dissections between GAMYB and Dof transcription factors suggest a role for protein-protein associations in the gibberellin-mediated expression of the RAmy1A gene in the rice aleurone. Plant Physiology, 133 (2):850-863.
pmid: 14500792 |
| [27] |
Wei P C, Tan F, Gao X Q, Zhang X Q, Wang G Q, Xu H, Li L J, Chen J, Wang X C. 2010. Overexpression of AtDOF4.7,an Arabidopsis DOF family transcription factor,induces floral organ abscission deficiency in Arabidopsis. Plant Physiol, 153 (3):1031-1045.
doi: 10.1104/pp.110.153247 URL |
| [28] |
Wei Q, Wang W, Hu T, Hu H, Mao W, Zhu Q, Bao C. 2018. Genome-wide identification and characterization of Dof transcription factors in eggplant (Solanum melongena L.). Peer J, 6:e4481.
doi: 10.7717/peerj.4481 URL |
| [29] |
Wen C L, Cheng Q, Zhao L, Mao A, Yang J, Yu S, Weng Y, Xu Y. 2016. Identification and characterisation of Dof transcription factors in the cucumber genome. Scientific Reports, 6:23072.
doi: 10.1038/srep23072 URL |
| [30] |
Wu Z, Cheng J, Cui J, Xu X, Liang G, Luo X, Chen X, Tang X, Hu K, Qin C. 2016. Genome-wide identification and expression profile of Dof transcription factor gene family in pepper(Capsicum annuum L.). Frontiers in Plant Science, 7:574.doi: 10.3389/fpls.2016.00574.
doi: 10.3389/fpls.2016.00574 |
| [31] | Xu P, Chen H, Cai W. 2020. Transcription factor CDF 4 promotes leaf senescence and floral organ abscission by regulating abscisic acid and reactive oxygen species pathways in Arabidopsis. EMBO Rep, 21 (7):e48967. |
| [32] |
Yanagisawa S. 1995. A novel DNA-binding domain that may form a single zinc finger motif. Nucleic Acids Research, 23 (17):3403-3410.
pmid: 7567449 |
| [33] |
Yanagisawa S. 2000. Dof1 and Dof 2 transcription factors are associated with expression of multiple genes involved in carbon metabolism in maize. The Plant Journal:for Cell and Molecular Biology, 21 (3):281-288.
doi: 10.1046/j.1365-313x.2000.00685.x URL |
| [34] |
Yanagisawa S, Izui K. 1993. Molecular cloning of two DNA-binding proteins of maize that are structurally different but interact with the same sequence motif. The Journal of Biological Chemistry, 268 (21):16028-16036.
doi: 10.1016/S0021-9258(18)82353-5 URL |
| [35] |
Yanagisawa S, Schmidt R J. 2010. Diversity and similarity among recognition sequences of Dof transcription factors. Plant Journal, 17 (2):209-214.
doi: 10.1046/j.1365-313X.1999.00363.x URL |
| [36] | Zhang Huanxin, Li Guoquan, Yang Huidong, Cao Na, Zhu Fanghong. 2019. Genome-wide identification and expression of Dof family in melon(Cucumis melo). Acta Horticulturae Sinica, 46 (11):2176-2187. (in Chinese) |
| 张焕欣, 李国权, 杨惠栋, 曹娜, 朱方红. 2019. 甜瓜Dof家族全基因组鉴定与表达分析. 园艺学报, 46 (11):2176-2187. | |
| [37] | Zhang Shaoling, Xie Zhihua. 2019. Current status,trends,main problems and the suggestions on development of pear industry in China. Journal of Fruit Science, 36 (8):1067-1072. (in Chinese) |
| 张绍铃, 谢智华. 2019. 我国梨产业发展现状、趋势、存在问题与对策建议. 果树学报, 36 (8):1067-1072. | |
| [38] |
Zhuo M, Sakuraba Y, Yanagisawa S A. 2020. Jasmonate-activated MYC2-Dof2.1-MYC 2 transcriptional loop promotes leaf senescence in Arabidopsis. Plant Cell., 32 (1):242-262.
doi: 10.1105/tpc.19.00297 URL |
| [1] | 宋健坤, 杨英杰, 李鼎立, 马春晖, 王彩虹, 王 然. 梨新品种‘鲁秀’[J]. 园艺学报, 2022, 49(S2): 3-4. |
| [2] | 董星光, 曹玉芬, 张 莹, 田路明, 霍宏亮, 齐 丹, 徐家玉, 刘 超, 王立东. 抗寒脆肉梨新品种‘玉翠香’[J]. 园艺学报, 2022, 49(S2): 5-6. |
| [3] | 欧春青, 姜淑苓, 王 斐, 马 力, 张艳杰, 刘振杰. 早熟梨新品种‘兴梨蜜水’[J]. 园艺学报, 2022, 49(S2): 7-8. |
| [4] | 张艳杰, 王 斐, 欧春青, 马 力, 姜淑苓, 刘振杰. 梨新品种‘中梨玉脆3’[J]. 园艺学报, 2022, 49(S2): 9-10. |
| [5] | 范 净, 陈启亮, 张靖国, 杨晓平, 杜 威, 田 瑞, 周德平, 胡红菊, . 中熟红皮砂梨新品种‘金彤’[J]. 园艺学报, 2022, 49(S2): 11-12. |
| [6] | 王苏珂, 李秀根, 杨 健, 王 龙, 苏艳丽, 张向展, 薛华柏. 红皮梨新品种‘丹霞红’[J]. 园艺学报, 2022, 49(S2): 13-14. |
| [7] | 王 斐, 欧春青, 张艳杰, 马 力, 姜淑苓. 晚熟耐贮梨新品种‘华秋’[J]. 园艺学报, 2022, 49(S1): 9-10. |
| [8] | 宋健坤, 李鼎立, 杨英杰, 马春晖, 王彩虹, 王 然. 梨新品种‘琴岛红’[J]. 园艺学报, 2022, 49(S1): 11-12. |
| [9] | 郭伟珍, 赵京献, 李莹. 中早熟梨新品种‘美玉’[J]. 园艺学报, 2022, 49(9): 2051-2052. |
| [10] | 陶鑫, 朱荣香, 贡鑫, 吴磊, 张绍铃, 赵建荣, 张虎平. 梨果糖激酶基因PpyFRK5在果实蔗糖积累中的作用[J]. 园艺学报, 2022, 49(7): 1429-1440. |
| [11] | 张秋悦, 刘昌来, 于晓晶, 杨甲定, 封超年. 盐胁迫条件下杜梨叶片差异表达基因qRT-PCR内参基因筛选[J]. 园艺学报, 2022, 49(7): 1557-1570. |
| [12] | 梁沁, 张延晖, 康开权, 刘瑾航, 李亮, 冯宇, 王超, 杨超, 李永裕. miR168家族进化特性及其在砂梨休眠期的表达模式分析[J]. 园艺学报, 2022, 49(5): 958-972. |
| [13] | 周徐子鑫, 杨威, 毛美琴, 薛彦斌, 马均. 金边红苞凤梨叶色突变体色素鉴定及类胡萝卜素合成限速基因筛选[J]. 园艺学报, 2022, 49(5): 1081-1091. |
| [14] | 向妙莲, 吴帆, 李树成, 马巧利, 王印宝, 肖刘华, 陈金印, 陈明. 外源褪黑素调控活性氧代谢诱导梨果实抗采后黑斑病[J]. 园艺学报, 2022, 49(5): 1102-1110. |
| [15] | 王宏, 杨王莉, 蔺经, 杨青松, 李晓刚, 盛宝龙, 常有宏. 早熟砂梨‘苏翠1号’与其亲本‘翠冠’‘华酥’成熟果实差异代谢产物及差异基因比较分析[J]. 园艺学报, 2022, 49(3): 493-508. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司