
园艺学报 ›› 2022, Vol. 49 ›› Issue (7): 1458-1472.doi: 10.16420/j.issn.0513-353x.2021-0351
蔡建法1,2, 莫雪莲2,3, 管思聪1,2, 陈栩2, 薛程1,*( )
)
收稿日期:2021-12-16
修回日期:2022-03-07
出版日期:2022-07-25
发布日期:2022-07-29
通讯作者:
薛程
E-mail:xcheng@sdau.edu.cn
基金资助:
CAI Jianfa1,2, MO Xuelian2,3, GUAN Sicong1,2, CHEN Xu2, XUE Cheng1,*( )
)
Received:2021-12-16
Revised:2022-03-07
Online:2022-07-25
Published:2022-07-29
Contact:
XUE Cheng
E-mail:xcheng@sdau.edu.cn
摘要:
在森林草莓中鉴定到5个YABBY家族基因,与其他蔷薇科植物进行系统发育树分析发现,YABBY家族成员被划分成5个亚族(INO、YAB2、YAB3、CRC、YAB5);各亚家族的外显子和内含子数量有所差异,但同一亚家族的数量相当;各蛋白基序之间离散程度差异较小,表明草莓YABBY家族成员生物学功能可能较为保守。组织特异性表达分析显示,FvYAB5.1在草莓匍匐茎的休眠芽与非休眠芽中表达水平差异显著。过量表达FvYAB5.1的转基因拟南芥叶片形态卷曲呈梭形,植株矮化,开花期推迟;茎尖分生组织(SAM)发育主效基因SHOOT MERISTEMLESS(STM)和KNAT2的表达水平受到显著抑制,生长素和细胞分裂素相关基因AtARF5、AtARR4、AtIPT7的转录表达水平明显升高,AtARR7和AtARR15的转录表达水平则受到了显著抑制。综合以上结果,推测FvYAB5.1可能通过调控细胞分裂素和生长素的拮抗关系,参与草莓匍匐茎节间SAM的发育过程,最终引起匍匐茎中休眠芽与非休眠芽的交替生长。
中图分类号:
蔡建法, 莫雪莲, 管思聪, 陈栩, 薛程. 草莓FvYABBY5.1表达特性和功能分析[J]. 园艺学报, 2022, 49(7): 1458-1472.
CAI Jianfa, MO Xuelian, GUAN Sicong, CHEN Xu, XUE Cheng. Expression Characteristics and Functional Analysis of FvYABBY5.1 in Strawberry[J]. Acta Horticulturae Sinica, 2022, 49(7): 1458-1472.
| 基因 Gene | 引物序列(5′-3′) Primer sequence |
|---|---|
| FvActin | F:ACCGTTGATTCGCACAATTGGTCATCG;R:TACTGCGGGTCGGCAATCGGACG |
| FvINO | F:GTCACTGCAGTTGCCTCCTA;R:TTTCCTGCGAACCCTGATCG |
| FvYAB2.1 | F:AATCCTGACATTAGCCATAGGG;R:CATGAGAAAATGACCATCGCTT |
| FvYAB3.1 | F:CTGCTATGTTCACTGCAACTTT;R:CATCATGTTAGGCTGATTGGTG |
| FvYAB3.2 | F:AAGTTTGTTCAAGACTGTGACG;R:TACTCCTCCTCTTGGTCTTACA |
| FvYAB5.1 | F:CTTTTGCAATATTGTTCTCGCG;R:CTTTGGATGATGAACCCGAATC |
| AtActin | F:GTCGTACAACCGGTATTGTGCTG;R:CTCTCTCTGTAAGGATCTTCATGAGGT |
| AtBP | F:TCATGGAAGCATACTGTGACA;R:TGACTCAGAAGGATATGGCCA |
| AtSTM | F:TGTCAGAAGGTTGGAGCACCA;R:TTTGTTGCTCCGAAGGGTAA |
| AtKNAT2 | F:GATTGCCAAAAGGTGGGAGC;R:TGTCGCCTTCAGTAGGGTA |
| AtARF5 | F:TTATCTTCTTGTAGCCTCTT;R:CCACACTCAACACTAACT |
| AtARR7 | F:TCAATGCCAGGACTTTCAGGA;R:TGCTCCTTCTTTGAGACATTCTTG |
| AtARR15 | F:TCAATGCCGGGACTAACAGG;R:TGCTCCTTCTATCATACATTGTTCT |
| AtARR4 | F:GACCAGAATCGACAGATGCCT;R:AGCTTCCGTTTGTTTCCGTT |
| AtIPT7 | F:GGAGGAAGTGGAAGTAGAG;R:CACACATACGAAATCTTGGA |
表1 qRT-PCR引物
Table 1 Primers for qRT-PCR
| 基因 Gene | 引物序列(5′-3′) Primer sequence |
|---|---|
| FvActin | F:ACCGTTGATTCGCACAATTGGTCATCG;R:TACTGCGGGTCGGCAATCGGACG |
| FvINO | F:GTCACTGCAGTTGCCTCCTA;R:TTTCCTGCGAACCCTGATCG |
| FvYAB2.1 | F:AATCCTGACATTAGCCATAGGG;R:CATGAGAAAATGACCATCGCTT |
| FvYAB3.1 | F:CTGCTATGTTCACTGCAACTTT;R:CATCATGTTAGGCTGATTGGTG |
| FvYAB3.2 | F:AAGTTTGTTCAAGACTGTGACG;R:TACTCCTCCTCTTGGTCTTACA |
| FvYAB5.1 | F:CTTTTGCAATATTGTTCTCGCG;R:CTTTGGATGATGAACCCGAATC |
| AtActin | F:GTCGTACAACCGGTATTGTGCTG;R:CTCTCTCTGTAAGGATCTTCATGAGGT |
| AtBP | F:TCATGGAAGCATACTGTGACA;R:TGACTCAGAAGGATATGGCCA |
| AtSTM | F:TGTCAGAAGGTTGGAGCACCA;R:TTTGTTGCTCCGAAGGGTAA |
| AtKNAT2 | F:GATTGCCAAAAGGTGGGAGC;R:TGTCGCCTTCAGTAGGGTA |
| AtARF5 | F:TTATCTTCTTGTAGCCTCTT;R:CCACACTCAACACTAACT |
| AtARR7 | F:TCAATGCCAGGACTTTCAGGA;R:TGCTCCTTCTTTGAGACATTCTTG |
| AtARR15 | F:TCAATGCCGGGACTAACAGG;R:TGCTCCTTCTATCATACATTGTTCT |
| AtARR4 | F:GACCAGAATCGACAGATGCCT;R:AGCTTCCGTTTGTTTCCGTT |
| AtIPT7 | F:GGAGGAAGTGGAAGTAGAG;R:CACACATACGAAATCTTGGA |
| 基因名称 Gene | 引物序列(5′-3′) Primer sequence |
|---|---|
| FvYAB5.1 | F:GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATGTCGAGCTGCATCGATG R:GGGGACCACTTTGTACAAGAAAGCTGGGTTAGCATAACTGGTGACAGGTACTG |
| FvYAB3.1 | F:GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATGTCTGCTGCTGCATCTT R:GGGGACCACTTTGTACAAGAAAGCTGGGTTGTAGGGGGAGACGCCC |
| FvYAB3.2 | F:GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATGTCATCCTCTAATTCTGCTTCTT R:GGGGACCACTTTGTACAAGAAAGCTGGGTTGTAAGGAGAGACCCGCACA |
表2 PCR引物
Table 2 Primers for PCR
| 基因名称 Gene | 引物序列(5′-3′) Primer sequence |
|---|---|
| FvYAB5.1 | F:GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATGTCGAGCTGCATCGATG R:GGGGACCACTTTGTACAAGAAAGCTGGGTTAGCATAACTGGTGACAGGTACTG |
| FvYAB3.1 | F:GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATGTCTGCTGCTGCATCTT R:GGGGACCACTTTGTACAAGAAAGCTGGGTTGTAGGGGGAGACGCCC |
| FvYAB3.2 | F:GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATGTCATCCTCTAATTCTGCTTCTT R:GGGGACCACTTTGTACAAGAAAGCTGGGTTGTAAGGAGAGACCCGCACA |
| 基因 Gene | 亚家族 Subfamily | 基因号 Gene ID | 染色体位置 Chromosome location | 蛋白长度/aa Protein length | 不稳定系数 Instability coefficient | 等电点 pI | 亲水性 Gravy |
|---|---|---|---|---|---|---|---|
| FvINO | INO | gene04115-v1.0-hybrid | scf0513158:3866711..3868355 | 237 | 46.40 | 5.51 | -0.659 |
| FvYAB3.1 | YAB3 | gene03938-v1.0-hybrid | scf0513153:189961..192371 | 218 | 47.85 | 8.24 | -0.312 |
| FvYAB2.1 | YAB2 | gene22887-v2.0.a2-hybrid | Chr03:14803114..14809010 | 447 | 62.07 | 10.01 | -0.503 |
| FvYAB5.1 | YAB5 | gene31056-v2.0.a2-hybrid | Chr01:1699533..1703162 | 180 | 45.54 | 8.19 | -0.422 |
| FvYAB3.2 | YAB3 | gene32251-v1.0-hybrid | scf0513098:214010..216501 | 232 | 45.90 | 8.58 | -0.424 |
表3 草莓植物YABBY家族鉴定和基本理化性质分析
Table3 Identification and analysis of basic physicochemical properties of YABBY family in strawberry
| 基因 Gene | 亚家族 Subfamily | 基因号 Gene ID | 染色体位置 Chromosome location | 蛋白长度/aa Protein length | 不稳定系数 Instability coefficient | 等电点 pI | 亲水性 Gravy |
|---|---|---|---|---|---|---|---|
| FvINO | INO | gene04115-v1.0-hybrid | scf0513158:3866711..3868355 | 237 | 46.40 | 5.51 | -0.659 |
| FvYAB3.1 | YAB3 | gene03938-v1.0-hybrid | scf0513153:189961..192371 | 218 | 47.85 | 8.24 | -0.312 |
| FvYAB2.1 | YAB2 | gene22887-v2.0.a2-hybrid | Chr03:14803114..14809010 | 447 | 62.07 | 10.01 | -0.503 |
| FvYAB5.1 | YAB5 | gene31056-v2.0.a2-hybrid | Chr01:1699533..1703162 | 180 | 45.54 | 8.19 | -0.422 |
| FvYAB3.2 | YAB3 | gene32251-v1.0-hybrid | scf0513098:214010..216501 | 232 | 45.90 | 8.58 | -0.424 |
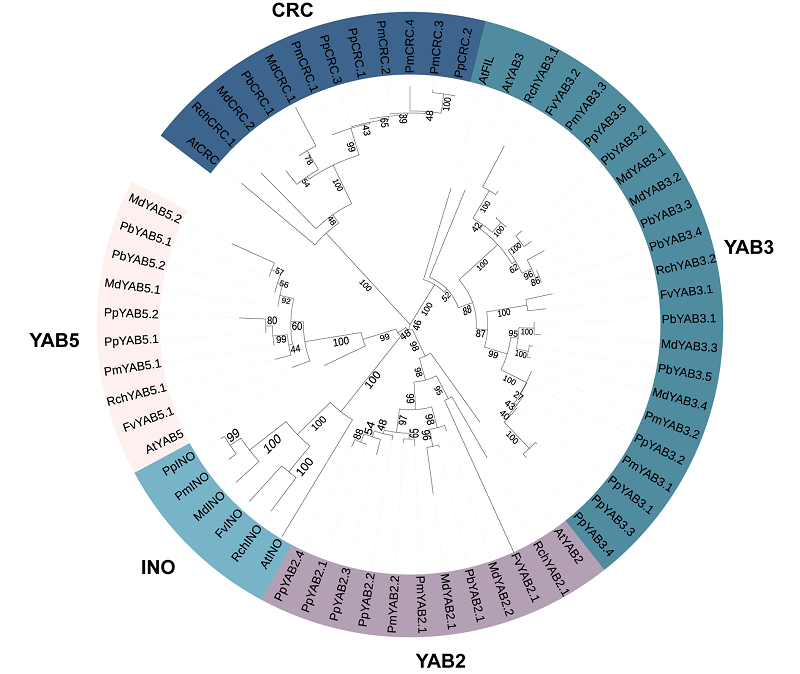
图1 拟南芥(At)、月季(Rch)、草莓(Fv)、梅(Pm)、桃(Pp)、梨(Pb)和苹果(Md)YABBY系统发育树
Fig. 1 Phylogenetic tree of YABBY proteins of Arabidopsis(At),Chinese rose(Rch),strawberry(Fv),plum(Pm),peach(Pp),pear(Pb)and apple(Md)
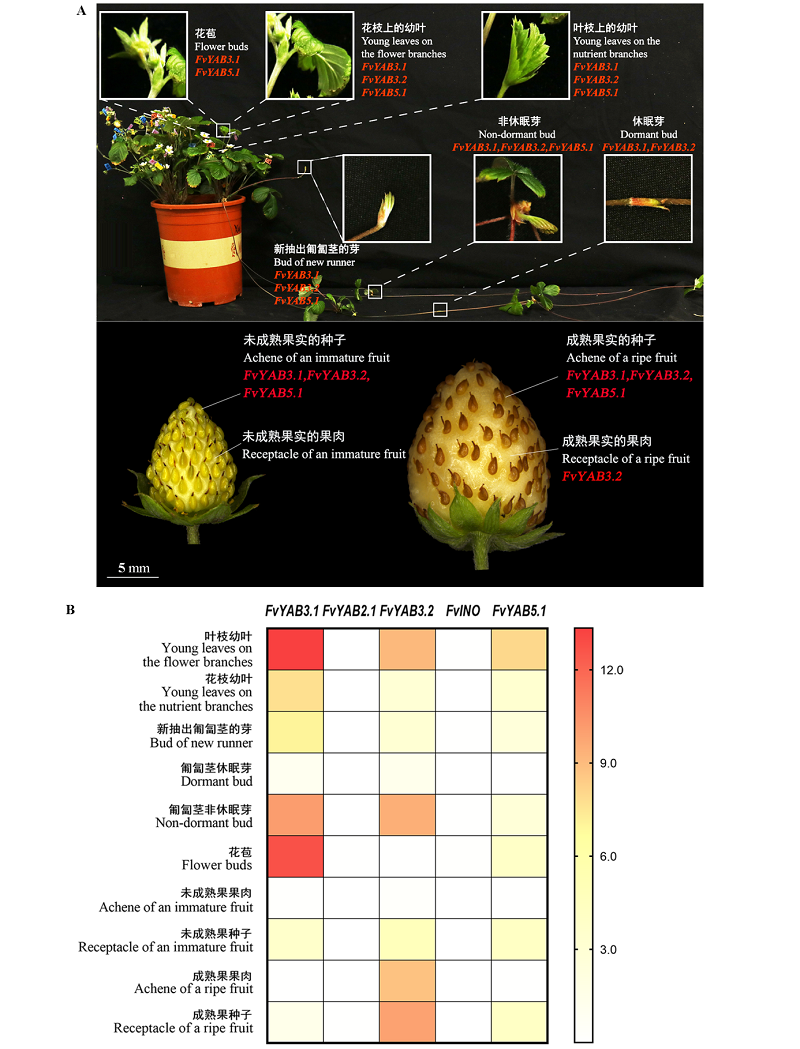
图4 ‘Hawaii-4’草莓各组织示意图(A)及YABBY家族成员组织特异性表达分析(B)
Fig. 4 Schematic drawing of various tissues of in Fragaria vesca‘Hawaii-4’(A)and specific expression analysis of YABBY family members in different organs(B)
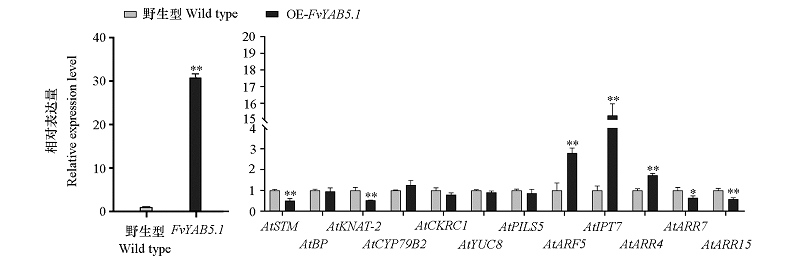
图8 茎尖分生组织生长发育、生长素和细胞分裂素相关基因在FvYAB5.1过表达(OE)拟南芥中的表达(播种后15 d)
Fig. 8 Transcript levels of shoot apical meristem,auxin and cytokinin related genes in 35S:FvYAB5.1-GFP transgenic Arabidopsis plants(15 days after sowing)t-test,* α = 0.05;** α = 0.01.
| [1] |
Alvarez J, Smyth D R. 1999. CRABS CLAW and SPATULA,two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development, 126 (11):2377-2386.
doi: 10.1242/dev.126.11.2377 pmid: 10225997 |
| [2] | Bailey T L, Boden M, Buske F A, Frith M, Grant C E, Clementi L, Ren J, Li W W, Noble W S. 2009. MEME SUITE:tools for motif discovery and searching. Nucleic Acids Research,37 (Web Server issue):W202-W208. |
| [3] |
Barbez E, Kubeš M, Rolčík J, Béziat C, Pěnčík A, Wang B, Rosquete MR, Zhu J, Dobrev PI, Lee Y, Zažímalovà E, Petrášek J, Geisler M, Friml J, Kleine-Vehn J. 2012. A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature, 485 (7396):119-222.
doi: 10.1038/nature11001 URL |
| [4] | Bartholmes C, Hidalgo O, Gleissberg S. 2012. Evolution of the YABBY gene family with emphasis on the basal eudicot Eschscholzia californica (Papaveraceae). Plant Biol(Stuttg), 14 (1):11-23. |
| [5] |
Bartley G E, Ishida B K. 2007. Ethylene-sensitive and insensitive regulation of transcription factor expression during in vitro tomato sepal ripening. J Exp Bot, 58 (8):2043-2051.
pmid: 17452748 |
| [6] |
Barton M, Poethig R. 1993. Formation of the shoot apical meristem in Arabidopsis thaliana:an analysis of development in the wild type and in the shoot meristemless mutant. Development, 119:823-831.
doi: 10.1242/dev.119.3.823 URL |
| [7] |
Boter M, Golz J F, Giménez-Ibañez S, Fernandez-Barbero G, Franco-Zorrilla J M, Solano R. 2015. FILAMENTOUS FLOWER is a direct target of JAZ3 and modulates responses to jasmonate. Plant Cell, 27 (11):3160-3174.
doi: 10.1105/tpc.15.00220 URL |
| [8] |
Bowman J L. 2000. The YABBY gene family and abaxial cell fate. Current Opinion in Plant Biology, 3 (1):17-22.
pmid: 10679447 |
| [9] |
Bowman J L, Eshed Y, Baum S F. 2002. Establishment of polarity in angiosperm lateral organs. Trends in Genetics, 18 (3):134-141.
pmid: 11858837 |
| [10] |
Bowman J L, Smyth D R. 1999. CRABS CLAW,a gene that regulates carpel and nectary development in Arabidopsis,encodes a novel protein with zinc finger and helix-loop-helix domains. Development, 126 (11):2387-2396.
doi: 10.1242/dev.126.11.2387 pmid: 10225998 |
| [11] |
Buechel S, Leibfried A, To J P C, Zhao Z, Andersen S U, Kieber J J, Lohmann J U. 2010. Role of A-type Arabidopsis response regulators in meristem maintenance and regeneration. European Journal of Cell Biology, 89 (2):279-284.
doi: 10.1016/j.ejcb.2009.11.016 URL |
| [12] |
Byrne M E, Barley R, Curtis M, Arroyo J M, Dunham M, Hudson A, Martienssen R A. 2000. Asymmetric leaves 1 mediates leaf patterning and stem cell function in Arabidopsis. Nature, 408 (6815):967-971.
doi: 10.1038/35050091 URL |
| [13] |
Chang L, Ramireddy E, Schmülling T. 2015. Cytokinin as a positional cue regulating lateral root spacing in Arabidopsis. J Exp Bot, 66 (15):4759-4768.
doi: 10.1093/jxb/erv252 pmid: 26019251 |
| [14] |
Chen C, Chen H, Zhang Y, Thomas H R, Frank M H, He Y, Xia R. 2020. TBtools:an integrative toolkit developed for interactive analyses of big biological data. Mol Plant, 13 (8):1194-1202.
doi: 10.1016/j.molp.2020.06.009 URL |
| [15] |
Ckurshumova W, Smirnova T, Marcos D, Zayed Y, Berleth T. 2014. Irrepressible MONOPTEROS/ARF5 promotes de novo shoot formation. New Phytol, 204 (3):556-566.
doi: 10.1111/nph.13014 pmid: 25274430 |
| [16] |
Cong B, Barrero L S, Tanksley S D. 2008. Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nature Genetics, 40 (6):800-804.
doi: 10.1038/ng.144 pmid: 18469814 |
| [17] |
Dai M, Hu Y, Zhao Y, Liu H, Zhou D X. 2007a. A WUSCHEL-LIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development. Plant Physiol, 144 (1):380-390.
doi: 10.1104/pp.107.095737 URL |
| [18] | Dai M, Zhao Y, Ma Q, Hu Y, Hedden P, Zhang Q, Zhou D X. 2007b. The rice YABBY1 gene is involved in the feedback regulation of gibberellin metabolism. Plant Physiol, 144 (1):121-133. |
| [19] | Deng Jiao, Su Mengyue, Liu Xuelian, Ou Kefang, Hu Zhengrong, Yang Pingfang. 2022. Transcriptome analysis revealed the formation mechanism of floral color of lotus‘Da Sajin’with bicolor petal. Acta Horticulturae Sinica, 49 (2):365-377. (in Chinese) |
| 邓娇, 苏梦月, 刘雪莲, 欧克芳, 户正荣, 杨平仿. 2022. 基于转录组分析揭示双色花莲‘大洒锦’花色形成机理. 园艺学报, 49 (2):365-377. | |
| [20] |
Di D W, Wu L, Zhang L, An C W, Zhang T Z, Luo P, Gao H H, Kriechbaumer V, Guo G Q. 2016. Functional roles of Arabidopsis CKRC2/YUCCA8 gene and the involvement of PIF4 in the regulation of auxin biosynthesis by cytokinin. Sci Rep, 6:36866..
doi: 10.1038/srep36866 URL |
| [21] |
Du F, Guan C, Jiao Y. 2018. Molecular mechanisms of leaf morphogenesis. Molecular Plant, 11 (9):1117-1134.
doi: 10.1016/j.molp.2018.06.006 URL |
| [22] |
Endrizzi K, Moussian B, Haecker A, Levin J Z, Laux T. 1996. The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J, 10 (6):967-979.
pmid: 9011081 |
| [23] | Filyushin M A, Slugin M A, Dzhos E A, Kochieva E Z, Shchennikova A V. 2018. Coexpression of YABBY1 and YABBY3 genes in lateral organs of tomato species(Solanum,Section Lycopersicon). Dokl Biochem Biophys, 478 (1):50-54. |
| [24] |
Finet C, Floyd S K, Conway S J, Zhong B, Scutt C P, Bowman J L. 2016. Evolution of the YABBY gene family in seed plants. Evol Dev, 18 (2):116-126.
doi: 10.1111/ede.12173 URL |
| [25] |
Garrido-Bigotes A, Torrejón M, Solano R, Figueroa C R. 2020. Interactions of JAZ repressors with anthocyanin biosynthesis-related transcription factors of Fragaria × ananassa. Agronomy, 10 (10). DOI: 10.3390/agronomy10101586.
doi: 10.3390/agronomy10101586 URL |
| [26] |
Golz J F, Roccaro M, Kuzoff R, Hudson A. 2004. GRAMINIFOLIA promotes growth and polarity of Antirrhinum leaves. Development, 131 (15):3661-3670.
doi: 10.1242/dev.01221 URL |
| [27] |
Hamant O, Nogué F, Belles-Boix E, Jublot D, Grandjean O, Traas J, Pautot V. 2002. The KNAT2 homeodomain protein interacts with ethylene and cytokinin signaling. Plant Physiol, 130 (2):657-665.
doi: 10.1104/pp.004564 URL |
| [28] |
Hu B, Jin J, Guo A Y, Zhang H, Luo J, Gao G. 2015. GSDS 2.0:an upgraded gene feature visualization server. Bioinformatics, 31 (8):1296-1297.
doi: 10.1093/bioinformatics/btu817 URL |
| [29] |
Huang Z, Van Houten J, Gonzalez G, Xiao H, van der Knaap E. 2013. Genome-wide identification,phylogeny and expression analysis of SUN,OFP and YABBY gene family in tomato. Molecular Genetics and Genomics, 288 (3):111-129.
doi: 10.1007/s00438-013-0733-0 URL |
| [30] |
Jung S, Lee T, Cheng C-H, Buble K, Zheng P, Yu J, Humann J, Ficklin S P, Gasic K, Scott K, Frank M, Ru S, Hough H, Evans K, Peace C, Olmstead M, DeVetter L W, McFerson J, Coe M, Wegrzyn J L, Staton M E, Abbott A G, Main D. 2019. 15 years of GDR:new data and functionality in the Genome Database for Rosaceae. Nucleic Acids Research, 47 (D1):D1137-D1145.
doi: 10.1093/nar/gky1000 URL |
| [31] |
Kanaya E, Nakajima N, Okada K. 2002. Non-sequence-specific DNA binding by the FILAMENTOUS FLOWER protein from Arabidopsis thaliana is reduced by EDTA. J Biol Chem, 277 (14):11957-11964.
doi: 10.1074/jbc.M108889200 pmid: 11812777 |
| [32] |
Kang C, Darwish O, Geretz A, Shahan R, Alkharouf N, Liu Z. 2013. Genome-scale transcriptomic insights into early-stage fruit development in woodland strawberry Fragaria vesca. The Plant Cell, 25 (6):1960-1978.
doi: 10.1105/tpc.113.111732 URL |
| [33] |
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X:molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35 (6):1547-1549.
doi: 10.1093/molbev/msy096 URL |
| [34] |
Larkin M A, Blackshields G, Brown N P, Chenna R, McGettigan P A, McWilliam H, Valentin F, Wallace I M, Wilm A, Lopez R, Thompson J D, Gibson T J, Higgins D G. 2007. Clustal W and Clustal X version 2.0. Bioinformatics, 23 (21):2947-2948.
doi: 10.1093/bioinformatics/btm404 pmid: 17846036 |
| [35] |
Letunic I, Bork P. 2019. Interactive tree of life(iTOL)v4:recent updates and new developments. Nucleic Acids Res, 47 (W1):W256-W259.
doi: 10.1093/nar/gkz239 |
| [36] | Liao Fanglei, Chen Zeyu, Xu Qiyue, Chen Wenrong, Guo Weidong. 2018. A review on fruit-shaping genes study:reference for fruit pattern formation in fingered citron. Acta Horticulturae Sinica, 45 (9):1701-1714. (in Chinese) |
| 廖芳蕾, 陈泽宇, 徐启越, 杨莉, 陈文荣, 郭卫东. 2018. 果形建成基因研究进展及其对佛手果形发育研究的启示. 园艺学报, 45 (9):1701-1714. | |
| [37] | Liao Fang-lei, Han Xiao-xia, Chen Wen-rong, Guo Yan, Zhang Chen-xiao, Chen Ze-yu, Zhou Ya-yan, Guo Wei-dong. 2016. Fruit morphogenesis observation and expression analysis of fruit-shaping-related genes in fingered citron. Acta Horticulturae Sinica, 43 (11):2141-2150. (in Chinese) |
| 廖芳蕾, 韩晓霞, 陈文荣, 郭艳, 张晨晓, 陈泽宇, 周亚艳, 郭卫东. 2016. 佛手果形发育观察及果形相关基因表达分析. 园艺学报, 43 (11):2141-2150. | |
| [38] |
Long J A, Moan E I, Medford J I, Barton M K. 1996. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature, 379 (6560):66-69.
doi: 10.1038/379066a0 URL |
| [39] |
Mele G, Ori N, Sato Y, Hake S. 2003. The knotted1-like homeobox gene BREVIPEDICELLUS regulates cell differentiation by modulating metabolic pathways. Genes Dev, 17 (17):2088-2093.
doi: 10.1101/gad.1120003 URL |
| [40] |
Müller D, Waldie T, Miyawaki K, To J P, Melnyk C W, Kieber J J, Kakimoto T, Leyser O. 2015. Cytokinin is required for escape but not release from auxin mediated apical dominance. Plant J, 82 (5):874-886.
doi: 10.1111/tpj.12862 URL |
| [41] |
Osakabe Y, Miyata S, Urao T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. 2002. Overexpression of Arabidopsis response regulators,ARR4/ATRR1/IBC7 and ARR8/ATRR3,alters cytokinin responses differentially in the shoot and in callus formation. Biochemical and Biophysical Research Communications, 293 (2):806-815.
pmid: 12054542 |
| [42] |
Pautot V, Dockx J, Hamant O, Kronenberger J, Grandjean O, Jublot D, Traas J. 2001. KNAT2:evidence for a link between knotted-like genes and carpel development. Plant Cell, 13 (8):1719-1734.
pmid: 11487688 |
| [43] |
Qiu Y, Guan S C, Wen C, Li P, Gao Z, Chen X. 2019. Auxin and cytokinin coordinate the dormancy and outgrowth of axillary bud in strawberry runner. BMC Plant Biol, 19 (1):528.
doi: 10.1186/s12870-019-2151-x URL |
| [44] | Rameau C, Bertheloot J, Leduc N, Andrieu B, Foucher F, Sakr S. 2014. Multiple pathways regulate shoot branching. Front Plant Sci, 5:741. |
| [45] |
Sarojam R, Sappl P G, Goldshmidt A, Efroni I, Floyd S K, Eshed Y, Bowman J L. 2010. Differentiating Arabidopsis shoots from leaves by combined YABBY activities. Plant Cell, 22 (7):2113-2130.
doi: 10.1105/tpc.110.075853 URL |
| [46] |
Schmid M, Davison T S, Henz S R, Pape U J, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann J U. 2005. A gene expression map of Arabidopsis thaliana development. Nat Genet, 37 (5):501-506.
doi: 10.1038/ng1543 URL |
| [47] | Shao Hongxia, Chen Pengfei, Zhang Dong, Wu Haiqin, Zhao Caiping, Han Mingyu. 2017. Identification,evolution and expression analysis of the YABBY gene family in apple(Malus × domestica Borkh.). Acta Agriculturae Zhejiangensis, 29 (7):1129-1138. (in Chinese) |
|
邵红霞, 陈鸿飞, 张东, 吴海芹, 赵彩平, 韩明玉. 2017. 苹果YABBY基因家族的鉴定、进化及表达分析. 浙江农业学报, 29 (7):1129-1138.
doi: 10.3969/j.issn.1004-1524.2017.07.10 |
|
| [48] |
Stahle M I, Kuehlich J, Staron L, von Arnim A G, Golz J F. 2009. YABBYs and the transcriptional corepressors LEUNIG and LEUNIG_HOMOLOG maintain leaf polarity and meristem activity in Arabidopsis. Plant Cell, 21 (10):3105-3118.
doi: 10.1105/tpc.109.070458 URL |
| [49] |
Toriba T, Harada K, Takamura A, Nakamura H, Ichikawa H, Suzaki T, Hirano H Y. 2007. Molecular characterization the YABBY gene family in Oryza sativa and expression analysis of OsYABBY1. Mol Genet Genomics, 277 (5):457-468.
pmid: 17216490 |
| [50] |
Villanueva J M, Broadhvest J, Hauser B A, Meister R J, Schneitz K, Gasser C S. 1999. INNER NO OUTER regulates abaxial- adaxial patterning in Arabidopsis ovules. Genes Dev, 13 (23):3160-3169.
doi: 10.1101/gad.13.23.3160 URL |
| [51] |
Waldie T, Leyser O. 2018. Cytokinin targets auxin transport to promote shoot branching. Plant Physiol, 177 (2):803-818.
doi: 10.1104/pp.17.01691 pmid: 29717021 |
| [52] | Wang A, Tang J, Li D, Chen C, Zhao X, Zhu L. 2009. Isolation and functional analysis of LiYAB1,a YABBY family gene,from lily(Lilium longiflorum). J Plant Physiol, 166 (9):988-995. |
| [53] |
Zhang S, Wang L, Sun X, Li Y, Yao J, van Nocker S, Wang X. 2019. Genome-wide analysis of the YABBY gene family in grapevine and functional characterization of VvYABBY4. Front Plant Sci, 10:1207.
doi: 10.3389/fpls.2019.01207 URL |
| [54] |
Zhang T, Li C, Li D, Liu Y, Yang X. 2020. Roles of YABBY transcription factors in the modulation of morphogenesis,development,and phytohormone and stress responses in plants. J Plant Res, 133 (6):751-763.
doi: 10.1007/s10265-020-01227-7 URL |
| [55] |
Zhang X, Henriques R, Lin S S, Niu Q W, Chua N H. 2006. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc, 1 (2):641-646.
doi: 10.1038/nprot.2006.97 URL |
| [56] |
Zhao W, Su H Y, Song J, Zhao X Y, Zhang X S. 2006. Ectopic expression of TaYAB1,a member of YABBY gene family in wheat,causes the partial abaxialization of the adaxial epidermises of leaves and arrests the development of shoot apical meristem in Arabidopsis. Plant Science, 170 (2):364-371.
doi: 10.1016/j.plantsci.2005.09.008 URL |
| [57] |
Zhao Y, Hull A K, Gupta N R, Goss K A, Alonso J, Ecker J R, Normanly J, Chory J, Celenza J L. 2002. Trp-dependent auxin biosynthesis in Arabidopsis:involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev, 16 (23):3100-12.
doi: 10.1101/gad.1035402 URL |
| [58] |
Zhao Z, Andersen S U, Ljung K, Dolezal K, Miotk A, Schultheiss S J, Lohmann J U. 2010. Hormonal control of the shoot stem-cell niche. Nature, 465 (7301):1089-1092.
doi: 10.1038/nature09126 URL |
| [1] | 宋艳红, 陈亚铎, 张晓玉, 宋 盼, 刘丽锋, 李 刚, 赵 霞, 周厚成, . 森林草莓FvbHLH130转录因子调控植株提前开花[J]. 园艺学报, 2023, 50(2): 295-306. |
| [2] | 何成勇, 赵晓丽, 许腾飞, 高德航, 李世访, 王红清. 草莓病毒1山东分离物全基因组分析[J]. 园艺学报, 2023, 50(1): 153-160. |
| [3] | 杨 雷, 李 莉, 董 辉, 冯 佳, 张建军, 范婧芳, 杨秋叶, 杨 莉, . 草莓新品种‘石莓11号’[J]. 园艺学报, 2022, 49(S2): 79-80. |
| [4] | 赵 霞, 李 刚, 刘丽锋, 宋艳红, 周厚成. 草莓新品种‘华硕1号’[J]. 园艺学报, 2022, 49(S2): 81-82. |
| [5] | 董 静, 常琳琳, 王桂霞, 钟传飞, 隗永青, 孙 健, 孙 瑞, 张宏力, 高用顺, 许利平, 陶 磅, 罗志伟, 张运涛, . 四季草莓新品种‘静红’[J]. 园艺学报, 2022, 49(S1): 61-62. |
| [6] | 杨丽媛, 王倩, 王许会, 徐通达, 马军. 草莓生长素合成关键酶FveTAA1保守氨基酸位点T111的生物学功能研究[J]. 园艺学报, 2021, 48(9): 1695-1705. |
| [7] | 洪燕红, 叶清华, 李泽坤, 王威, 谢倩, 陈清西, 陈建清. 红花草莓‘莓红’花瓣花色苷积累及其MYB基因的表达分析[J]. 园艺学报, 2021, 48(8): 1470-1484. |
| [8] | 张志强, 卢世雄, 马宗桓, 李彦彪, 高彩霞, 陈佰鸿, 毛娟. 草莓LIM家族候选基因的鉴定及在非生物胁迫下的表达[J]. 园艺学报, 2021, 48(8): 1485-1503. |
| [9] | 黄倩茹, 牛永浩, 吴宽, 刘占杰, 曹孟籍, 赵磊, 吴云锋. 陕西省草莓病毒种类的LncRNA测序初步鉴定[J]. 园艺学报, 2021, 48(8): 1589-1594. |
| [10] | 谷思, 刘璐, 李安然, 张伟伟, 赵帅琪, 邢宇. 草莓果实酵母双杂交文库的构建及FvM4K1互作蛋白的筛选[J]. 园艺学报, 2021, 48(6): 1067-1078. |
| [11] | 黄威剑, 李梦. 果树全基因组测序现状与展望[J]. 园艺学报, 2021, 48(4): 733-748. |
| [12] | 齐永志, 孙雅如, 王冰, 郭邯菲, 马可, 甄文超. 草莓根系分泌物和腐解物化感作用研究进展[J]. 园艺学报, 2021, 48(4): 778-790. |
| [13] | 刘璐, 谷思, 张伟伟, 赵帅琪, 李安然, 邢宇. 森林草莓FvMOB1与FvM4K1互作及响应生长素调控初探[J]. 园艺学报, 2021, 48(11): 2185-2196. |
| [14] | 陈道, 张洁, 吴祖建, 丁新伦. 草莓白化相关病毒中国分离物全基因组分析[J]. 园艺学报, 2021, 48(1): 146-152. |
| [15] | 罗 贺, 李伟佳, 李 贺, 张志宏. 草莓FaRGA1基因沉默改变开花和匍匐茎抽生特性[J]. 园艺学报, 2020, 47(12): 2331-2339. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司