
园艺学报 ›› 2022, Vol. 49 ›› Issue (6): 1213-1232.doi: 10.16420/j.issn.0513-353x.2021-0331
张凯, 麻明英, 王萍, 李益, 金燕, 盛玲, 邓子牛, 马先锋( )
)
收稿日期:2021-12-14
修回日期:2022-02-25
出版日期:2022-06-25
发布日期:2022-07-04
通讯作者:
马先锋
E-mail:ma8006@hunau.edu.cn
基金资助:
ZHANG Kai, MA Mingying, WANG Ping, LI Yi, JIN Yan, SHENG Ling, DENG Ziniu, MA Xianfeng( )
)
Received:2021-12-14
Revised:2022-02-25
Online:2022-06-25
Published:2022-07-04
Contact:
MA Xianfeng
E-mail:ma8006@hunau.edu.cn
摘要:
为探究HSP20在柑橘(Citrus L.)响应溃疡病菌(Xanthomonas citri subsp. citri,Xcc)侵染过程中发挥的作用,鉴定CsHSP20家族基因。生物学信息分析显示共有42个,分为11个亚族,分布在9条染色体上,编码135 ~ 373个氨基酸,蛋白分子量15.17 ~ 41.71 kD,等电点4.53 ~ 10.07,92.9%的基因没有或只有1个内含子,各亚族基因间有高度相似的结构和保守基序,每个基因的启动子都具有与激素或胁迫相关的顺式作用元件。其次,对感病的冰糖橙和抗病的枸橼C-05进行接种Xcc和40 ℃热胁迫处理,qRT-PCR结果显示CsHSP20家族中的HSP17.9、HSP23.3、HSP18.5和HSP18.0均上调表达,但抗感材料之间差异明显,HSP23.3在枸橼C-05中受Xcc上调表达水平极显著,该结果与受Xcc侵染的转录组结果一致。在冰糖橙叶片瞬时超表达候选基因,Xcc定量分析结果显示枸橼C-05的HSP23.3抑制Xcc繁殖速度更明显。
中图分类号:
张凯, 麻明英, 王萍, 李益, 金燕, 盛玲, 邓子牛, 马先锋. 柑橘HSP20家族基因鉴定及其响应溃疡病菌侵染表达分析[J]. 园艺学报, 2022, 49(6): 1213-1232.
ZHANG Kai, MA Mingying, WANG Ping, LI Yi, JIN Yan, SHENG Ling, DENG Ziniu, MA Xianfeng. Identification of HSP20 Family Genes in Citrus and Their Expression in Pathogen Infection Responses Citrus Canker[J]. Acta Horticulturae Sinica, 2022, 49(6): 1213-1232.
| 目的基因引物 Primer | 上游引物序列(5′-3′) Forward primer sequence | 下游引物序列(5′-3′) Reverse primer sequence |
|---|---|---|
| EF-1α(qRT-PCR) | GTAACCAAGTCTGCTGCCAAG | GACCCAAACACCCAACACATT |
| CsHSP17.9(qRT-PCR) | TGGTACGAAAGCAAACCCCT | TCAGACTTACAATTGCAAACACACA |
| CmHSP17.9(qRT-PCR) | TGGGCCATAGAAGAAGCAGC | AGCATCCGTAACGACACCAA |
| CsHSP23.3(qRT-PCR) | CTTCCGGGTGCTAGAGCAAA | CGTTTCCTCTCTCCGCTCAC |
| CsHSP18.5(qRT-PCR) | TTTTCCATCTGCGGTGTCCT | CCTTAGCCCCGGAAGATCAG |
| CsHSP18.0(qRT-PCR) | AAGCATTTCCGAAGTTGTTGAG | TTGGGATGTTTCCCTAGCCG |
| CmHSP18.0(qRT-PCR) | TGGGCCATAGAAGAAGCAGC | AGCATCCGTAACGACACCAA |
| CsHSP17.9(ORF) | TAGGTACCATGTCACTCATTCCAAGCATCT | TAGGTACCGCCAGAGATCTCAATAGCCTTA |
| CmHSP17.9(ORF) | GGGAGCTCATGTCACTCATTCCAAGCATCT | AAGAGCTCAGAGCTGGCATCTGTTCCAGG |
| CsHSP23.3(ORF) | TAGGTACCATGAGGCAGCAACAACAAACA | TAGGTACCAAGCTCTTGCCTAGCCTCGCTC |
| CmHSP23.3(ORF) | TAGGTACCATGAGACAGCAACAACGAACAC | AAGGTACCAAGCTCTTGCCTAGCCTCGCTC |
| CsHSP18.5(ORF) | TAGGTACCATGTCGCTGATTCCAAGCTTCT | TAGGTACCGCCAGAGATTTGAATAGCCCTG |
表1 本研究中使用的引物序列
Table 1 Primer sequences used in this study
| 目的基因引物 Primer | 上游引物序列(5′-3′) Forward primer sequence | 下游引物序列(5′-3′) Reverse primer sequence |
|---|---|---|
| EF-1α(qRT-PCR) | GTAACCAAGTCTGCTGCCAAG | GACCCAAACACCCAACACATT |
| CsHSP17.9(qRT-PCR) | TGGTACGAAAGCAAACCCCT | TCAGACTTACAATTGCAAACACACA |
| CmHSP17.9(qRT-PCR) | TGGGCCATAGAAGAAGCAGC | AGCATCCGTAACGACACCAA |
| CsHSP23.3(qRT-PCR) | CTTCCGGGTGCTAGAGCAAA | CGTTTCCTCTCTCCGCTCAC |
| CsHSP18.5(qRT-PCR) | TTTTCCATCTGCGGTGTCCT | CCTTAGCCCCGGAAGATCAG |
| CsHSP18.0(qRT-PCR) | AAGCATTTCCGAAGTTGTTGAG | TTGGGATGTTTCCCTAGCCG |
| CmHSP18.0(qRT-PCR) | TGGGCCATAGAAGAAGCAGC | AGCATCCGTAACGACACCAA |
| CsHSP17.9(ORF) | TAGGTACCATGTCACTCATTCCAAGCATCT | TAGGTACCGCCAGAGATCTCAATAGCCTTA |
| CmHSP17.9(ORF) | GGGAGCTCATGTCACTCATTCCAAGCATCT | AAGAGCTCAGAGCTGGCATCTGTTCCAGG |
| CsHSP23.3(ORF) | TAGGTACCATGAGGCAGCAACAACAAACA | TAGGTACCAAGCTCTTGCCTAGCCTCGCTC |
| CmHSP23.3(ORF) | TAGGTACCATGAGACAGCAACAACGAACAC | AAGGTACCAAGCTCTTGCCTAGCCTCGCTC |
| CsHSP18.5(ORF) | TAGGTACCATGTCGCTGATTCCAAGCTTCT | TAGGTACCGCCAGAGATTTGAATAGCCCTG |
| 基因名称 Gene name | CAP登录号 CAP ID | 染色体 Chr. | 基因位置 Genomic position | 开放阅读框 ORF | 外显子数 Exon number | 氨基酸数 Amino acid number | 分子量/ kD MW | 等电点 pI |
|---|---|---|---|---|---|---|---|---|
| CsHSP20-01 | Cs1g12560 | 1 | 15649712 ~ 15650511 (-) | 800 | 2 | 235 | 26.81 | 9.08 |
| CsHSP20-02 | Cs1g16780 | 1 | 20054644 ~ 20057349 (-) | 539 | 3 | 135 | 15.17 | 9.01 |
| CsHSP20-03 | Cs2g07920 | 2 | 4751770 ~ 4753509 (-) | 490 | 3 | 136 | 15.44 | 4.94 |
| CsHSP20-04 | Cs2g24040 | 2 | 22865474 ~ 22866152 (+) | 417 | 1 | 138 | 15.65 | 5.79 |
| CsHSP20-05 | Cs2g24360 | 2 | 23415680 ~ 23417091 (-) | 865 | 2 | 229 | 25.80 | 6.77 |
| CsHSP20-06 | Cs2g28910 | 2 | 28489066 ~ 28490856 (+) | 1 630 | 2 | 308 | 34.20 | 9.26 |
| CsHSP20-07 | Cs3g07660 | 3 | 10958719 ~ 10959189 (-) | 471 | 1 | 156 | 18.08 | 5.70 |
| CsHSP20-08 | Cs3g13530 | 3 | 17951675 ~ 17954257 (+) | 2 280 | 3 | 208 | 22.15 | 4.81 |
| CsHSP20-09 | Cs3g21390 | 3 | 24314830 ~ 24316680 (-) | 1 232 | 2 | 373 | 41.71 | 4.80 |
| CsHSP20-10 | Cs4g04260 | 4 | 2440105 ~ 2443442 (-) | 453 | 1 | 150 | 16.84 | 4.53 |
| CsHSP20-11 | Cs4g05780 | 4 | 3542671 ~ 3543153 (-) | 483 | 1 | 160 | 17.87 | 5.57 |
| CsHSP20-12 | Cs4g05800 | 4 | 3545945 ~ 3546616 (-) | 489 | 1 | 162 | 18.39 | 6.45 |
| CsHSP20-13 | Cs4g05880 | 4 | 3592334 ~ 3593451 (-) | 477 | 1 | 158 | 17.80 | 5.37 |
| CsHSP20-14 | Cs4g12260 | 4 | 9326064 ~ 9327838 (+) | 773 | 2 | 198 | 22.48 | 5.48 |
| CsHSP20-15 | Cs5g04230 | 5 | 2361326 ~ 2362866 (+) | 819 | 2 | 180 | 20.34 | 4.73 |
| CsHSP20-16 | Cs5g18500 | 5 | 18849146 ~ 18850530 (+) | 612 | 1 | 203 | 23.26 | 5.77 |
| CsHSP20-17 | Cs5g26930 | 5 | 29657216 ~ 29657893 (+) | 630 | 2 | 182 | 20.96 | 10.07 |
| CsHSP20-18 | Cs5g26940 | 5 | 29661034 ~ 29661937 (+) | 715 | 2 | 207 | 23.08 | 9.10 |
| CsHSP20-19 | Cs6g07310 | 6 | 9164497 ~ 9165164 (-) | 471 | 1 | 156 | 17.78 | 7.93 |
| CsHSP20-20 | Cs6g07320 | 6 | 9166603 ~ 9167820 (+) | 438 | 1 | 145 | 16.34 | 7.92 |
| CsHSP20-21 | Cs6g17370 | 6 | 17879116 ~ 17880307 (-) | 1 192 | 2 | 148 | 16.55 | 6.92 |
| CsHSP20-22 | Cs7g10040 | 7 | 6437870 ~ 6438966 (-) | 779 | 2 | 224 | 25.41 | 8.67 |
| CsHSP20-23 | Cs7g23870 | 7 | 22776659 ~ 22778441 (-) | 1 597 | 2 | 339 | 37.75 | 5.30 |
| CsHSP20-24 | Cs7g23890 | 7 | 22813654 ~ 22814811 (-) | 1 062 | 2 | 309 | 34.41 | 8.20 |
| CsHSP20-25 | Cs7g32260 | 7 | 31935895 ~ 31936951 (-) | 471 | 1 | 156 | 17.64 | 5.55 |
| CsHSP20-26 | Cs8g13310 | 8 | 16029454 ~ 16030281 (+) | 828 | 1 | 275 | 30.11 | 5.68 |
| CsHSP20-27 | Cs8g13370 | 8 | 16132947 ~ 16133375 (+) | 429 | 1 | 142 | 15.91 | 5.84 |
| CsHSP20-28 | Cs8g13450 | 8 | 16211348 ~ 16212457 (+) | 1 110 | 1 | 369 | 40.91 | 5.97 |
| CsHSP20-29 | Cs8g13500 | 8 | 16261568 ~ 16262230 (+) | 663 | 1 | 220 | 23.89 | 8.02 |
| CsHSP20-30 | Cs8g13560 | 8 | 16334995 ~ 16335974 (+) | 980 | 1 | 224 | 24.62 | 5.75 |
| CsHSP20-31 | Cs8g18020 | 8 | 20763153 ~ 20764015 (+) | 477 | 1 | 158 | 17.98 | 5.84 |
| CsHSP20-32 | Cs8g18360 | 8 | 20993803 ~ 20994946 (-) | 477 | 1 | 158 | 17.89 | 6.77 |
| CsHSP20-33 | Cs8g19490 | 8 | 21656820 ~ 21657642 (-) | 486 | 1 | 161 | 18.28 | 6.17 |
| CsHSP20-34 | Cs8g19510 | 8 | 21667991 ~ 21668854 (-) | 486 | 1 | 161 | 18.22 | 6.76 |
| CsHSP20-35 | Cs8g19520 | 8 | 21670200 ~ 21671498 (+) | 489 | 1 | 162 | 18.47 | 5.79 |
| CsHSP20-36 | Cs8g19530 | 8 | 21674289 ~ 21675349 (+) | 492 | 1 | 163 | 18.46 | 6.76 |
| CsHSP20-37 | Cs8g19540 | 8 | 21677280 ~ 21677985 (-) | 453 | 1 | 150 | 17.15 | 6.18 |
| CsHSP20-38 | Cs9g14690 | 9 | 14093289 ~ 14096486 (-) | 1 036 | 2 | 207 | 23.37 | 5.43 |
| CsHSP20-39 | orange1.1t01849 | Un | 29239328 ~ 29242931 (+) | 939 | 1 | 312 | 34.78 | 8.56 |
| CsHSP20-40 | orange1.1t03235 | Un | 49911150 ~ 49912086 (+) | 489 | 1 | 162 | 17.98 | 7.13 |
| CsHSP20-41 | orange1.1t03877 | Un | 59580055 ~ 59580573 (-) | 519 | 1 | 172 | 19.78 | 7.93 |
| CsHSP20-42 | orange1.1t03881 | Un | 59667559 ~ 59668167 (-) | 609 | 1 | 202 | 23.10 | 6.61 |
表2 CsHSP20家族基因特征
Table 2 Features of HSP20 family genes in Citrus sinensis
| 基因名称 Gene name | CAP登录号 CAP ID | 染色体 Chr. | 基因位置 Genomic position | 开放阅读框 ORF | 外显子数 Exon number | 氨基酸数 Amino acid number | 分子量/ kD MW | 等电点 pI |
|---|---|---|---|---|---|---|---|---|
| CsHSP20-01 | Cs1g12560 | 1 | 15649712 ~ 15650511 (-) | 800 | 2 | 235 | 26.81 | 9.08 |
| CsHSP20-02 | Cs1g16780 | 1 | 20054644 ~ 20057349 (-) | 539 | 3 | 135 | 15.17 | 9.01 |
| CsHSP20-03 | Cs2g07920 | 2 | 4751770 ~ 4753509 (-) | 490 | 3 | 136 | 15.44 | 4.94 |
| CsHSP20-04 | Cs2g24040 | 2 | 22865474 ~ 22866152 (+) | 417 | 1 | 138 | 15.65 | 5.79 |
| CsHSP20-05 | Cs2g24360 | 2 | 23415680 ~ 23417091 (-) | 865 | 2 | 229 | 25.80 | 6.77 |
| CsHSP20-06 | Cs2g28910 | 2 | 28489066 ~ 28490856 (+) | 1 630 | 2 | 308 | 34.20 | 9.26 |
| CsHSP20-07 | Cs3g07660 | 3 | 10958719 ~ 10959189 (-) | 471 | 1 | 156 | 18.08 | 5.70 |
| CsHSP20-08 | Cs3g13530 | 3 | 17951675 ~ 17954257 (+) | 2 280 | 3 | 208 | 22.15 | 4.81 |
| CsHSP20-09 | Cs3g21390 | 3 | 24314830 ~ 24316680 (-) | 1 232 | 2 | 373 | 41.71 | 4.80 |
| CsHSP20-10 | Cs4g04260 | 4 | 2440105 ~ 2443442 (-) | 453 | 1 | 150 | 16.84 | 4.53 |
| CsHSP20-11 | Cs4g05780 | 4 | 3542671 ~ 3543153 (-) | 483 | 1 | 160 | 17.87 | 5.57 |
| CsHSP20-12 | Cs4g05800 | 4 | 3545945 ~ 3546616 (-) | 489 | 1 | 162 | 18.39 | 6.45 |
| CsHSP20-13 | Cs4g05880 | 4 | 3592334 ~ 3593451 (-) | 477 | 1 | 158 | 17.80 | 5.37 |
| CsHSP20-14 | Cs4g12260 | 4 | 9326064 ~ 9327838 (+) | 773 | 2 | 198 | 22.48 | 5.48 |
| CsHSP20-15 | Cs5g04230 | 5 | 2361326 ~ 2362866 (+) | 819 | 2 | 180 | 20.34 | 4.73 |
| CsHSP20-16 | Cs5g18500 | 5 | 18849146 ~ 18850530 (+) | 612 | 1 | 203 | 23.26 | 5.77 |
| CsHSP20-17 | Cs5g26930 | 5 | 29657216 ~ 29657893 (+) | 630 | 2 | 182 | 20.96 | 10.07 |
| CsHSP20-18 | Cs5g26940 | 5 | 29661034 ~ 29661937 (+) | 715 | 2 | 207 | 23.08 | 9.10 |
| CsHSP20-19 | Cs6g07310 | 6 | 9164497 ~ 9165164 (-) | 471 | 1 | 156 | 17.78 | 7.93 |
| CsHSP20-20 | Cs6g07320 | 6 | 9166603 ~ 9167820 (+) | 438 | 1 | 145 | 16.34 | 7.92 |
| CsHSP20-21 | Cs6g17370 | 6 | 17879116 ~ 17880307 (-) | 1 192 | 2 | 148 | 16.55 | 6.92 |
| CsHSP20-22 | Cs7g10040 | 7 | 6437870 ~ 6438966 (-) | 779 | 2 | 224 | 25.41 | 8.67 |
| CsHSP20-23 | Cs7g23870 | 7 | 22776659 ~ 22778441 (-) | 1 597 | 2 | 339 | 37.75 | 5.30 |
| CsHSP20-24 | Cs7g23890 | 7 | 22813654 ~ 22814811 (-) | 1 062 | 2 | 309 | 34.41 | 8.20 |
| CsHSP20-25 | Cs7g32260 | 7 | 31935895 ~ 31936951 (-) | 471 | 1 | 156 | 17.64 | 5.55 |
| CsHSP20-26 | Cs8g13310 | 8 | 16029454 ~ 16030281 (+) | 828 | 1 | 275 | 30.11 | 5.68 |
| CsHSP20-27 | Cs8g13370 | 8 | 16132947 ~ 16133375 (+) | 429 | 1 | 142 | 15.91 | 5.84 |
| CsHSP20-28 | Cs8g13450 | 8 | 16211348 ~ 16212457 (+) | 1 110 | 1 | 369 | 40.91 | 5.97 |
| CsHSP20-29 | Cs8g13500 | 8 | 16261568 ~ 16262230 (+) | 663 | 1 | 220 | 23.89 | 8.02 |
| CsHSP20-30 | Cs8g13560 | 8 | 16334995 ~ 16335974 (+) | 980 | 1 | 224 | 24.62 | 5.75 |
| CsHSP20-31 | Cs8g18020 | 8 | 20763153 ~ 20764015 (+) | 477 | 1 | 158 | 17.98 | 5.84 |
| CsHSP20-32 | Cs8g18360 | 8 | 20993803 ~ 20994946 (-) | 477 | 1 | 158 | 17.89 | 6.77 |
| CsHSP20-33 | Cs8g19490 | 8 | 21656820 ~ 21657642 (-) | 486 | 1 | 161 | 18.28 | 6.17 |
| CsHSP20-34 | Cs8g19510 | 8 | 21667991 ~ 21668854 (-) | 486 | 1 | 161 | 18.22 | 6.76 |
| CsHSP20-35 | Cs8g19520 | 8 | 21670200 ~ 21671498 (+) | 489 | 1 | 162 | 18.47 | 5.79 |
| CsHSP20-36 | Cs8g19530 | 8 | 21674289 ~ 21675349 (+) | 492 | 1 | 163 | 18.46 | 6.76 |
| CsHSP20-37 | Cs8g19540 | 8 | 21677280 ~ 21677985 (-) | 453 | 1 | 150 | 17.15 | 6.18 |
| CsHSP20-38 | Cs9g14690 | 9 | 14093289 ~ 14096486 (-) | 1 036 | 2 | 207 | 23.37 | 5.43 |
| CsHSP20-39 | orange1.1t01849 | Un | 29239328 ~ 29242931 (+) | 939 | 1 | 312 | 34.78 | 8.56 |
| CsHSP20-40 | orange1.1t03235 | Un | 49911150 ~ 49912086 (+) | 489 | 1 | 162 | 17.98 | 7.13 |
| CsHSP20-41 | orange1.1t03877 | Un | 59580055 ~ 59580573 (-) | 519 | 1 | 172 | 19.78 | 7.93 |
| CsHSP20-42 | orange1.1t03881 | Un | 59667559 ~ 59668167 (-) | 609 | 1 | 202 | 23.10 | 6.61 |
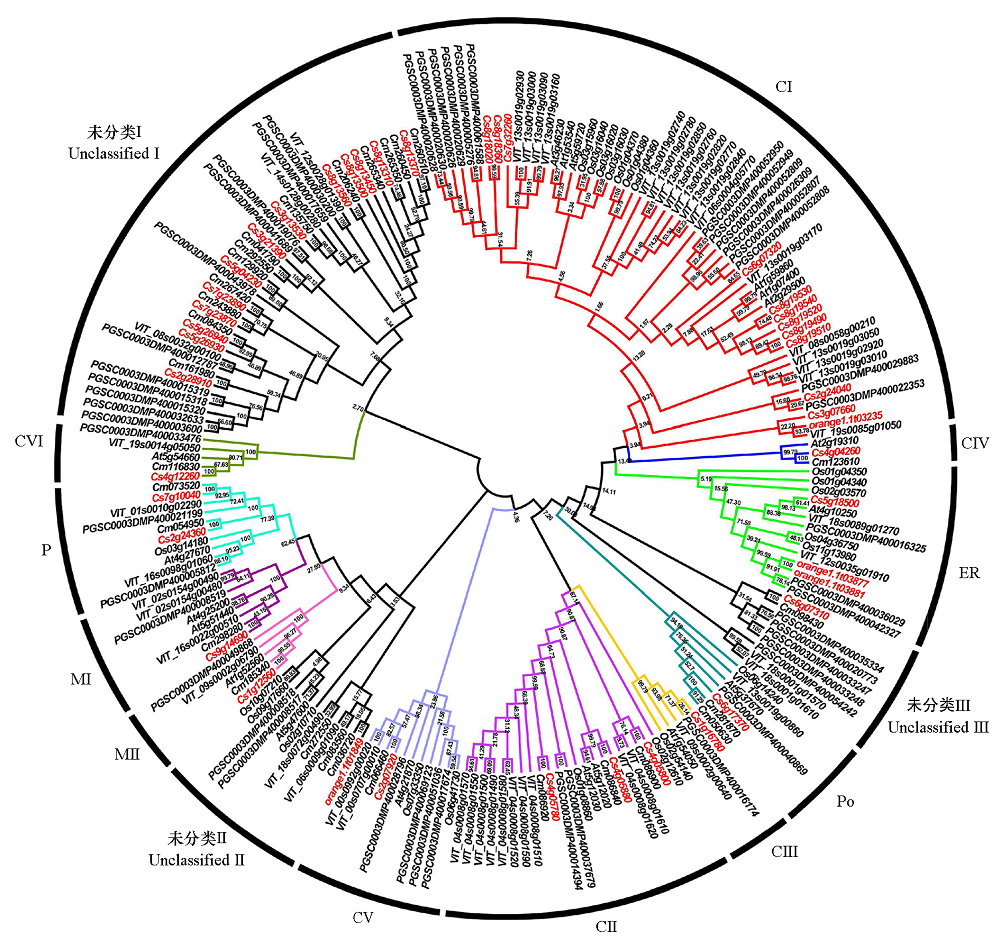
图2 不同物种HSP20基因的系统进化关系 Cs:甜橙;Cm:枸橼C-05;At:拟南芥;Os:水稻;PGSC:马铃薯;VIT:葡萄。进化分支上的数字为自展值(%),值越大表示进化分支置信度越高。
Fig. 2 The phylogenetic relationships of HSP20 genes in different species Cs:C. sinensis;Cm:C. medica;At:Arabidopsis thaliana;Os:Oryza sativa;PGSC:Solanum tuberosum;VIT:Vitis vinifera. The numbers on the branch are bootstrap value(%),the larger the value,the higher the confidence of the evolutionary branch.
| 保守基序名称 Motif name | 氨基酸序列 Amino acid sequence | 起始位点 Site | 序列长度/aa Width |
|---|---|---|---|
| Motif 1 | WKETPEAHVFKADLPGLRKEEVKVE | 32 | 25 |
| Motif 2 | KMDQIKASMENGVLTVTVPK | 33 | 20 |
| Motif 3 | SSGMFSRRFRLPENA | 31 | 15 |
| Motif 4 | VEDDRVLQISGQRKIEKEDKN | 27 | 21 |
| Motif 5 | MSLIPSFFGNRRSSVFDPFSLDVWDPFRDF | 8 | 30 |
| Motif 6 | DTWHRVER | 16 | 8 |
| Motif 7 | LSFLKKNEGDTTSPIGPMEFNLPAGLNTNDFETAIDDEGLLTLTFKKLVP | 5 | 50 |
| Motif 8 | MCLGNNPYQCSVFWYEDVNLVHFKYPLPPGAKQEDVKVEID | 7 | 41 |
| Motif 9 | EEEKKPEVKAIEISG | 11 | 15 |
| Motif 10 | QFPQETSAFVNTRVD | 9 | 15 |
表3 主要序列构建的保守结构基序
Table 3 Conserved structural motifs built from the primary sequences
| 保守基序名称 Motif name | 氨基酸序列 Amino acid sequence | 起始位点 Site | 序列长度/aa Width |
|---|---|---|---|
| Motif 1 | WKETPEAHVFKADLPGLRKEEVKVE | 32 | 25 |
| Motif 2 | KMDQIKASMENGVLTVTVPK | 33 | 20 |
| Motif 3 | SSGMFSRRFRLPENA | 31 | 15 |
| Motif 4 | VEDDRVLQISGQRKIEKEDKN | 27 | 21 |
| Motif 5 | MSLIPSFFGNRRSSVFDPFSLDVWDPFRDF | 8 | 30 |
| Motif 6 | DTWHRVER | 16 | 8 |
| Motif 7 | LSFLKKNEGDTTSPIGPMEFNLPAGLNTNDFETAIDDEGLLTLTFKKLVP | 5 | 50 |
| Motif 8 | MCLGNNPYQCSVFWYEDVNLVHFKYPLPPGAKQEDVKVEID | 7 | 41 |
| Motif 9 | EEEKKPEVKAIEISG | 11 | 15 |
| Motif 10 | QFPQETSAFVNTRVD | 9 | 15 |
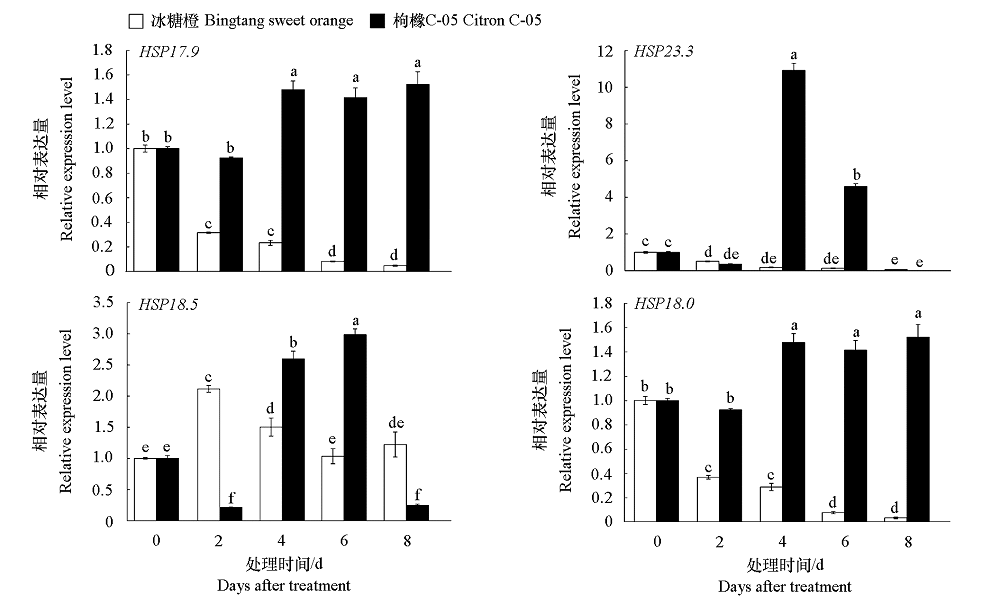
图5 HSP20候选基因受105 CFU · mL-1 Xcc的表达分析 不同字母代表基因表达量有显著性差异(P < 0.05)。下同。
Fig. 5 Expression analysis of HSP20 candidate genes in response to 105 CFU · mL-1 Xcc Different letters indicate significant differences(P < 0.05). The same below.

图8 瞬时超表达HSP20候选基因冰糖橙叶片症状 A:缓冲液对照;B:HSP20超表达载体;C:缓冲液 + Xcc;D:超表达载体 + Xcc。1:CsHSP23.3;2:CmHSP23.3;3:CsHSP17.9;4:CmHSP17.9;5:CsHSP18.5;6:CmHSP18.5;P:空载体对照。
Fig. 8 Symptoms in resistance identification of sweet orange leaves A:Buffer control;B:HSP20 candidate genes overexpression vector Agrobacterium;C:Buffer + Xcc;D:HSP20 candidate genes overexpression vector agrobacterium + Xcc。1:CsHSP23.3;2:CmHSP23.3;3:CsHSP17.9;4:CmHSP17.9;5:CsHSP18.5;6:CmHSP18.5;P:pCAMIBA1300 vector control.
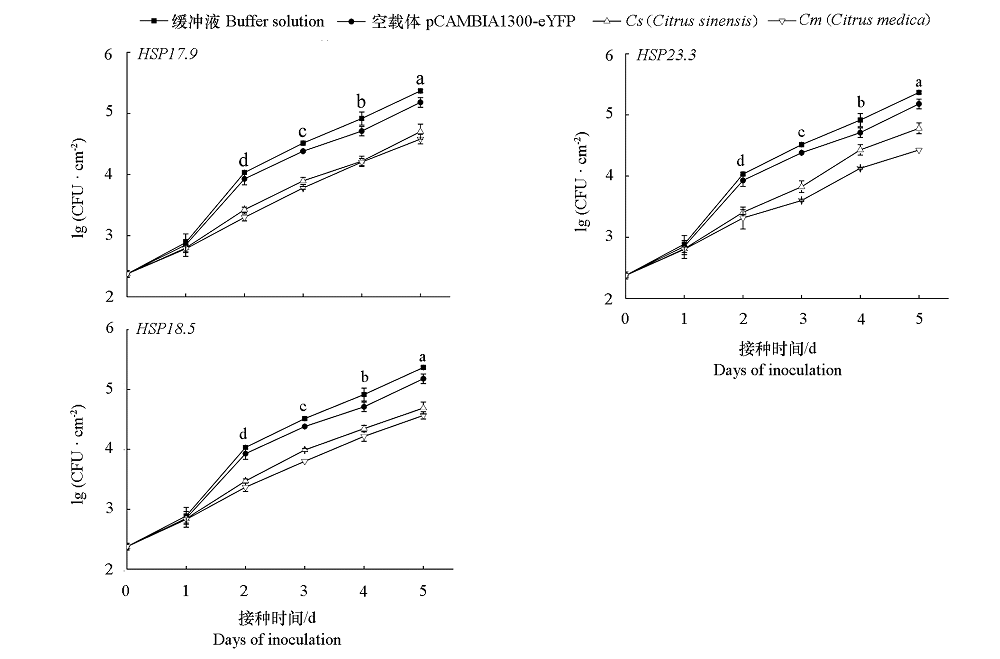
图9 冰糖橙叶片超表达HSP20候选基因的Xcc定量分析 不同字母代表缓冲液处理和超表达HSP20候选基因同一天Xcc生长量[单位面积Xcc含量,lg(CFU · cm-2)]有显著性差异(P < 0.05)。
Fig. 9 Xcc quantitative analysis of overexpression of HSP20 candidate genes in sweet orange leaves Different letters indicate significant difference at 0.05 levels Xcc growth by buffer and overexpression of HSP20 candidate genes on the same day.
| [1] | Alshameri A, Al-Qurainy F, Gaafar A-R, Khan S, Nadeem M, Alansi S. 2020. Identification of heat-responsive genes in guar[Cyamopsis tetragonoloba(L.)Taub]. International Journal of Genomics, 2020:3126592. |
| [2] |
Basha E, O’Neill H, Vierling E. 2012. Small heat shock proteins and alpha-crystallins:dynamic proteins with flexible functions. Trends in Biochemical Sciences, 37 (3):106-117.
doi: 10.1016/j.tibs.2011.11.005 pmid: 22177323 |
| [3] |
Behlau F, Gochez A M, Jones J B. 2020. Diversity and copper resistance of Xanthomonas affecting citrus. Tropical Plant Pathology, 45:200-212.
doi: 10.1007/s40858-020-00340-1 URL |
| [4] |
Bhaskar P B, Venkateshwaran M, Wu L, Ane J M, Jiang J. 2009. Agrobacterium-mediated transient gene expression and silencing:a rapid tool for functional gene assay in potato. PLoS ONE, 4 (6):e5812.
doi: 10.1371/journal.pone.0005812 URL |
| [5] |
Bondino H G, Valle E M, Have A T. 2012. Evolution and functional diversification of the small heat shock protein/α-crystallin family in higher plants. Planta, 235 (6):1299-1313.
doi: 10.1007/s00425-011-1575-9 URL |
| [6] |
Boter M, Amigues B, Peart J, Breuer C, Kadota Y, Casais C, Moore G, Kleanthous C, Ochsenbein F, Shirasu K, Guerois R. 2007. Structural and functional analysis of SGT 1 reveals that its interaction with HSP90 is required for the accumulation of Rx,an R protein involved in plant immunity. Plant Cell, 19 (11):3791-3804.
doi: 10.1105/tpc.107.050427 URL |
| [7] | Canto T. 2016. Transient expression systems in plants:potentialities and constraints. Adv Exp Med Biol, 896:287-301. |
| [8] |
Deng Z N, Xu L, Li D Z, Long G Y, Liu L P, Fang F, Shu G P. 2010. Screening citrus genotypes for resistance to canker disease(Xanthomonas axonopodis pv. citri). Plant Breeding, 129 (3):341-345.
doi: 10.1111/j.1439-0523.2009.01695.x URL |
| [9] |
Eisenhardt B D. 2013. Small heat shock proteins:recent developments. Biomol Concepts, 4 (6):583-595.
doi: 10.1515/bmc-2013-0028 pmid: 25436758 |
| [10] |
Escandon M, Valledor L, Pascual J, Pinto G, Canal M J, Meijon M. 2017. System-wide analysis of short-term response to high temperature in Pinus radiata. Journal of Experimental Botany, 68 (13):3629-3641.
doi: 10.1093/jxb/erx198 URL |
| [11] |
Ference C M, Gochez A M, Behlau F, Wang N, Graham J H, Jones J B. 2018. Recent advances in the understanding of Xanthomonas citri ssp. citri pathogenesis and citrus canker disease management. Molecular Plant Pathology, 19 (6):1302-1318.
doi: 10.1111/mpp.12638 URL |
| [12] | Figueiredo J F, Romer P, Lahaye T, Graham J H, White F F, Jones J B. 2011. Agrobacterium-mediated transient expression in citrus leaves:a rapid tool for gene expression and functional gene assay. Plant Cell Report, 30 (7):1339-1345. |
| [13] |
Fu H, Zhao M, Xu J, Tan L, Han J, Li D, Wang M, Xiao S, Ma X, Deng Z. 2020. Citron C-05 inhibits both the penetration and colonization of Xanthomonas citri subsp. citri to achieve resistance to citrus canker disease. Horticulture Research, 7:58-70.
doi: 10.1038/s41438-020-0278-4 URL |
| [14] |
Gallegos J E, Rose A B. 2019. An intron-derived motif strongly increases gene expression from transcribed sequences through a splicing independent mechanism in Arabidopsis thaliana. Scientific Reports, 9 (1):13777.
doi: 10.1038/s41598-019-50389-5 pmid: 31551463 |
| [15] |
Gochez A M, Behlau F, Singh R, Ong K, Whilby L, Jones J B. 2020. Panorama of citrus canker in the United States. Tropical Plant Pathology, 45:192-199.
doi: 10.1007/s40858-020-00355-8 URL |
| [16] |
Gottwald T R, Graham J H, Schubert T S. 2002. Citrus canker:the pathogen and its impact. Plant Health Progress, 3 (1):15-49.
doi: 10.1094/PHP-2002-0812-01-RV URL |
| [17] |
Graham J H, Gottwald T R, Cubero J, Achor D S. 2004. Xanthomonas axonopodis pv. citri:factors affecting successful eradication of citrus canker. Molecular Plant Pathology, 5 (1):1-15.
doi: 10.1046/j.1364-3703.2004.00197.x pmid: 20565577 |
| [18] |
Green B J, Fujiki M, Mett V, Kaczmarczyk J, Shamloul M, Musiychuk K, Underkoffler S, Yusibov V, Mett V. 2009. Transient protein expression in three Pisum sativum(green pea)varieties. Biotechnol J, 4 (2):230-237.
doi: 10.1002/biot.200800256 URL |
| [19] | Guo M, Liu J, Lu J, Zhai Y, Wang H, Gong Z, Wang S, Lu M. 2015. Genome-wide analysis of the CaHsp20 gene family in pepper:comprehensive sequence and expression profile analysis under heat stress. Frontiers in Plant Science, 6:806-812. |
| [20] |
Haq S U, Khan A, Ali M, Khattak A M, Gai W, Zhang H, Wei A, Gong Z. 2019. Heat shock proteins:dynamic biomolecules to counter plant biotic and abiotic stresses. International Journal of Molecular Sciences, 20 (21):5321-5352.
doi: 10.3390/ijms20215321 URL |
| [21] |
Haslbeck M, Vierling E. 2015. A first line of stress defense:small heat shock proteins and their function in protein homeostasis. Journal of Molecular Biology, 427 (7):1537-1548.
doi: 10.1016/j.jmb.2015.02.002 pmid: 25681016 |
| [22] |
Hu T, Sun X, Zhang X, Nevo E, Fu J. 2014a. An RNA sequencing transcriptome analysis of the high-temperature stressed tall fescue reveals novel insights into plant thermotolerance. BMC Genomics 15:1147-1160.
doi: 10.1186/1471-2164-15-1147 URL |
| [23] | Hu Y, Zhang J, Jia H, Sosso D, Li T, Frommer W B, Yang B, White F F, Wang N, Jones J B. 2014b. Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc Natl Acad Sci, 111 (4):E521-E529. |
| [24] |
Jacob P, Hirt H, Bendahmane A. 2017. The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnology Journal, 15 (4):405-414.
doi: 10.1111/pbi.12659 URL |
| [25] |
Ji X, Yu Y, Ni P, Zhang G, Guo D. 2019. Genome-wide identification of small heat-shock protein(HSP20)gene family in grape and expression profile during berry development. BMC Plant Biology, 19 (1):433-448.
doi: 10.1186/s12870-019-2031-4 URL |
| [26] |
Joensuu J J, Conley A J, Lienemann M, Brandle J E, Linder M B, Menassa R. 2010. Hydrophobin fusions for high-level transient protein expression and purification in Nicotiana benthamiana. Plant Physiol, 152 (2):622-633.
doi: 10.1104/pp.109.149021 pmid: 20018596 |
| [27] |
Jones H D, Doherty A, Sparks C A. 2009. Transient transformation of plants. Methods Mol Biol, 513:131-152.
doi: 10.1007/978-1-59745-427-8_8 pmid: 19347644 |
| [28] |
Ju Y, Tian H, Zhang R, Zuo L, Jin G, Xu Q, Ding X, Li X, Chu Z. 2017. Overexpression of OsHSP18.0-CI enhances resistance to bacterial leaf streak in rice. Rice, 10 (1):12-23.
doi: 10.1186/s12284-017-0153-6 URL |
| [29] |
Kim J H, Lim S D, Jang C S. 2019. Oryza sativa heat-induced RING finger protein 1(OsHIRP1)positively regulates plant response to heat stress. Plant Molecular Biology, 99 (6):545-559.
doi: 10.1007/s11103-019-00835-9 URL |
| [30] |
Kotak S, Larkindale J, Lee U, Koskull-Doring P V, Vierling E, Scharf K. 2007. Complexity of the heat stress response in plants. Current Opinion in Plant Biology, 10 (3):310-316.
doi: 10.1016/j.pbi.2007.04.011 URL |
| [31] |
Krenek P, Samajova O, Luptovciak I, Doskocilova A, Komis G, Samaj J. 2015. Transient plant transformation mediated by Agrobacterium tumefaciens:Principles,methods and applications. Biotechnology Advances, 33 (6 Pt 2):1024-1042.
doi: 10.1016/j.biotechadv.2015.03.012 pmid: 25819757 |
| [32] |
Kuang J, Liu J, Mei J, Wang C, Hu H, Zhang Y, Sun M, Ning X, Xiao L, Yang L. 2017. A Class II small heat shock protein OsHsp18.0 plays positive roles in both biotic and abiotic defense responses in rice. Sci Rep, 7 (1):11333.
doi: 10.1038/s41598-017-11882-x pmid: 28900229 |
| [33] |
Li J, Xiang C Y, Yang J, Chen J P, Zhang H M. 2015. Interaction of HSP 20 with a viral RdRp changes its sub-cellular localization and distribution pattern in plants. Scientific Reports, 5:14016.
doi: 10.1038/srep14016 URL |
| [34] |
Li J, Zhang H, Hu J, Liu J, Liu K. 2012. A heat shock protein gene,CsHsp45.9,involved in the response to diverse stresses in cucumber. Biochemical Genetic, 50 (7-8):565-578.
doi: 10.1007/s10528-012-9501-9 URL |
| [35] | Li Qiang, Qi Jingjing, Dou Wanfu, Qin Xiujuan, He Yongrui, Chen Shanchun. 2020. Overexpression of CsNBS-LRR in citrus confers bacterial canker resistance by regulating SA signaling pathway. Acta Horticulturae Sinica, 47 (5):817-826. (in Chinese) |
| 李强, 祁静静, 窦万福, 秦秀娟, 何永睿, 陈善春. 2020. 柑橘超量表达CsNBS-LRR通过SA信号途径增强对溃疡病抗性. 园艺学报 47 (5):817-826. | |
| [36] | Long Qin, Xie Yu, Lei Tiangang, He Yongrui, Xu Lanzhen, Zou Xiuping, Chen Shanchun. 2020a. Bioinformatics analysis of MAPK genes in citrus and their expression in response to canker disease. Acta Horticulturae Sinica, 47 (11):2095-2106. (in Chinese) |
| 龙琴, 谢宇, 雷天刚, 何永睿, 许兰珍, 邹修平, 陈善春. 2020a. 柑橘MAPK基因的生物信息及其响应溃疡病菌侵染的表达分析. 园艺学报, 47 (11):2095-2106. | |
| [37] | Long Qin, Xie Yu, Xu Lanzhen, He Yongrui, Zou Xiuping, Chen Shanchun. 2020b. Characteristics and mechanism of programmed cell death in response to citrus canker pathogen in the early stage of infection. Acta Horticulturae Sinica, 47 (6):1047-1058. (in Chinese) |
| 龙琴, 谢宇, 许兰珍, 何永睿, 邹修平, 陈善春. 2020b. 溃疡病菌侵染早期柑橘细胞程序性死亡的响应特征及机制. 园艺学报, 47 (6):1047-1058. | |
| [38] |
Lopes-Caitar V S, Carvalho M C D, Darben L M, Kuwahara M K, Nepomuceno A L, Dias W P, Abdelnoor R V, Marcelino-Guimarães F C. 2013. Genome-wide analysis of the Hsp20gene family in soybean:comprehensive sequence,genomic organization and expression profile analysis under abiotic and biotic stresses. BMC Genomics, 14:577-594.
doi: 10.1186/1471-2164-14-577 pmid: 23985061 |
| [39] |
Lozano R, Hamblin M T, Prochnik S, Jannink J L. 2015. Identification and distribution of the NBS-LRR gene family in the Cassava genome. BMC Genomics, 16:360-374.
doi: 10.1186/s12864-015-1554-9 URL |
| [40] |
MacRae T H. 2000. Structure and function of small heat shock/α-crystallin proteins:established concepts and emerging ideas. Cellular and Molecular Life Sciences, 57:899-913.
pmid: 10950306 |
| [41] |
Mu C, Zhang S, Yu G, Chen N, Li X, Liu H. 2013. Overexpression of small heat shock protein LimHSP16.45 in Arabidopsis enhances tolerance to abiotic stresses. PLoS ONE, 8 (12):e82264.
doi: 10.1371/journal.pone.0082264 URL |
| [42] | Ong K, Alabi O J. 2016. Xanthomonas citri(citrus canker). Extension Plant Pathology. |
| [43] |
Ouyang Y, Chen J, Xie W, Wang L, Zhang Q. 2009. Comprehensive sequence and expression profile analysis of Hsp20 gene family in rice. Plant Molecular Biology, 70 (3):341-357.
doi: 10.1007/s11103-009-9477-y pmid: 19277876 |
| [44] | Park C, Seo Y. 2015. Heat shock proteins:a review of the molecular chaperones for plant immunity. Plant Pathology Journal, 31 (4):323-333. |
| [45] |
Pei Q Y, Yu T, Wu T, Yang Q H, Gong K, Zhou R, Cui C L, Yu Y, Zhao W, Kang X, Cao R, Song X M. 2021. Comprehensive identification and analyses of the Hsf gene family in the whole-genome of three Apiaceae species. Horticultural Plant Journal, 7 (5):457-458.
doi: 10.1016/j.hpj.2020.08.005 URL |
| [46] |
Qi W Q, Zhang C L, Wang W J, Cao Z, Li S, Li H, Zhu W, HuangY Q, Bao M Z, HeY H, Zheng R R. 2021. Comparative transcriptome analysis of different heat stress responses between self-root grafting line and heterogeneous grafting line in rose. Horticultural Plant Journal, 7 (3):243-255.
doi: 10.1016/j.hpj.2021.03.004 URL |
| [47] |
Sarkar N K, Kim Y, Grover A. 2009. Rice sHsp genes:genomic organization and expression profiling under stress and development. BMC Genomics, 10:393-411.
doi: 10.1186/1471-2164-10-393 URL |
| [48] |
Schaad N W, Postnikova E, Lacy G, Sechler A, Agarkova I, Stromberg P E, Stromberg V K, Vidaver A K. 2006. Emended classification of xanthomonad pathogens on citrus. Systematic and Applied Microbiology, 29 (8):690-695.
doi: 10.1016/j.syapm.2006.08.001 URL |
| [49] | Scharf K, Siddique M, Vierling E. 2001. The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing α-crystallin domains(Acd proteins). Cell Stress & Chaperones, 6 (3):225-237. |
| [50] |
Sewelam N, Kazan K, Hudig M, Maurino V G, Schenk P M. 2019. The AtHSP17.4C1 gene expression is mediated by diverse signals that link biotic and abiotic stress factors with ROS and can be a useful molecular marker for oxidative stress. International Journal of Molecular Sciences, 20 (13):3201-3218.
doi: 10.3390/ijms20133201 URL |
| [51] | Shiotani H, Ozaki K, Tsuyumu S. 2000. Pathogenic interactions between Xanthomonas axonopodis pv. citri and cultivars of pummelo(Citrus grandis). Phytopathology, 91 (12):1383-1389. |
| [52] |
Siddique M, Gernhard S, Koskull-Döring P V, Vierling E, Scharf K. 2008. The plant sHSP superfamily:five new members in Arabidopsis thaliana with unexpected properties. Cell Stress & Chaperones, 13 (2):183-197.
doi: 10.1007/s12192-008-0032-6 URL |
| [53] |
Sumares J A P, Morão L G, Martins P M M, Martins D A B, Gomes E, Jr J B, Ferreira H. 2016. Temperature stress promotes cell division arrest in Xanthomonas citri subsp. citri. Microbiologyopen, 5 (2):244-253.
doi: 10.1002/mbo3.323 pmid: 26663580 |
| [54] | Sun W, Montagu M V, Verbruggen N. 2002. Small heat shock proteins and stress tolerance in plants. Biochimica et Biophysica Acta, 1577:1-9. |
| [55] |
Vernière C J, Gottwald T R, Pruvost O. 2003. Disease Development and symptom expression of Xanthomonas axonopodis pv. citri in various citrus plant tissues. Phytopathology, 93 (7):832-843.
doi: 10.1094/PHYTO.2003.93.7.832 pmid: 18943164 |
| [56] | Wang J, Chen D, Lei Y, Chang J W, Hao B H, Xing F, Li S, Xu Q, Deng X X, Chen L L. 2014. Citrus sinensis annotation project(CAP):a comprehensive database for sweet orange genome. PLoS ONE, 9 (1):e87723. |
| [57] | Wang Jing, Tan Fangjun, Liang Chengliang, Zhang Xilu, Ou Lijun, Niran Juntawong, Wang Fei, Jiao Chunhai, Zou Xuexiao, Chen Wenchao. 2020. Genome-wide identification and analysis of HSP 90 gene family in pepper. Acta Horticulturae Sinica, 47 (4):665-674. (in Chinese) |
| 王静, 谭放军, 梁成亮, 张西露, 欧立军, Niran Juntawong, 王飞, 焦春海, 邹学校, 陈文超. 2020. 辣椒热激蛋白HSP90家族基因鉴定及分析. 园艺学报, 47 (4):665-674. | |
| [58] |
Wang W, Vinocur B, Shoseyov O, Altman A. 2004. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. TRENDS in Plant Science, 9 (5):244-252.
doi: 10.1016/j.tplants.2004.03.006 URL |
| [59] |
Waters E R. 2013. The evolution,function,structure,and expression of the plant sHSPs. Journal of Experimental Botany, 64 (2):391-403.
doi: 10.1093/jxb/ers355 URL |
| [60] | Xu Jing, Tan Limei, Fu Hongyan, Zhu Zhimei, Long Libing, Hu Zhe, Ma Xianfeng, Deng Ziniu. 2021. Genetic diversity analysis of 14 citron genotypes based on molecular markers. Molecular Plant Breeding, 19 (14):4726-4737. (in Chinese) |
| 徐静, 谭李梅, 符红艳, 朱志媚, 龙栎冰, 胡哲, 马先锋, 邓子牛. 2021. 利用分子标记对14份枸橼种质进行多样性分析. 分子植物育种, 19 (14):4726-4737. | |
| [61] |
Xu Q, Chen L L, Ruan X, Chen D, Zhu A, Chen C, Bertrand D, Jiao W B, Hao B H, Lyon M P, Chen J, Gao S, Xing F, Lan H, Chang J W, Ge X, Lei Y, Hu Q, Miao Y, Wang L, Xiao S, Biswas M K, Zeng W, Guo F, Cao H, Yang X, Xu X W, Cheng Y J, Xu J, Liu J H, Luo O J, Tang Z, Guo W W, Kuang H, Zhang H Y, Roose M L, Nagarajan N, Deng X X, Ruan Y. 2013. The draft genome of sweet orange(Citrus sinensis). Nature Genetics, 45 (1):59-66.
doi: 10.1038/ng.2472 URL |
| [62] |
Yang M, Zhang Y, Zhang H, Wang H, Wei T, Che S, Zhang L, Hu B, Long H, Song W, Yu W, Yan G. 2017. Identification of MsHsp20 gene family in malus sieversii and functional characterization of MsHsp16.9 in heat tolerance. Frontiers in Plant Science, 8:1761-1778.
doi: 10.3389/fpls.2017.01761 URL |
| [63] | Yao Tingshan, Zhou Yan, Zhou Changyong. 2015. Research development of the differentiation and control of citrus bacterial canker disease. Acta Horticulturae Sinica, 42 (9):1699-1706. (in Chinese) |
| 姚廷山, 周彦, 周常勇. 2015. 柑橘溃疡病菌分化及防治研究进展. 园艺学报, 42 (9):1699-1706. | |
| [64] |
Ye S, Yu S, Shu L, Wu J, Wu A, Luo L. 2012. Expression profile analysis of 9 heat shock protein genes throughout the life cycle and under abiotic stress in rice. Chinese Science Bulletin, 57 (4):336-343.
doi: 10.1007/s11434-011-4863-7 URL |
| [65] | Yu J, Cheng Y, Feng K, Li Z, Ruan M, Ye Q, Wang R, Zhou G, Yao Z, Yang Y, Wan H. 2016. Genome-wide identification and expression profiling of tomato Hsp20 gene family in response to biotic and abiotic stresses. Frontiers in Plant Science, 7:1215-1229. |
| [66] |
Zhao P, Wang D, Wang R, Kong N, Zhang C, Yang C, Wu W, Ma H, Chen Q. 2018. Genome-wide analysis of the potato Hsp20gene family:identification,genomic organization and expression profiles in response to heat stress. BMC Genomics, 19 (1):61-74.
doi: 10.1186/s12864-018-4443-1 URL |
| [67] | Zhao Xi, Zhang Tingting, Xing Wenting, Wang Jian, Song Xiqiang, Zhou Yang. 2021. Genome-wide identification and expression analysis under temperature stress of HSP 70 gene family in Dendrobium catenatum. Acta Horticulturae Sinica, 48 (9):1743-1754. (in Chinese) |
| 赵溪, 张婷婷, 邢文婷, 王健, 宋希强, 周扬. 2021. 铁皮石斛热激蛋白HSP70基因家族鉴定及温度胁迫下的表达分析. 园艺学报, 48 (9):1743-1754. | |
| [68] | Zhu Zhimei, Tan Limei, Xu Jing, Fu Hongyan, Hu Zhe, Gong Lei, Yang Guibing, Wang Ping, Ma Xianfeng, Deng Ziniu. 2021. Evaluation of citron genotypes for the resistance to citrus canker. China Fruits,(1):43-49,110-111. (in Chinese) |
| 朱志媚, 谭李梅, 徐静, 符红艳, 胡哲, 龚蕾, 杨贵兵, 王萍, 马先锋, 邓子牛. 2021. 枸橼种质对柑橘溃疡病的抗性鉴定. 中国果树,(1):43-49,110-111. |
| [1] | 叶子茂, 申晚霞, 刘梦雨, 王 彤, 张晓楠, 余 歆, 刘小丰, 赵晓春, . R2R3-MYB转录因子CitMYB21对柑橘类黄酮生物合成的影响[J]. 园艺学报, 2023, 50(2): 250-264. |
| [2] | 蒋靖东, 韦壮敏, 王楠, 朱晨桥, 叶俊丽, 谢宗周, 邓秀新, 柴利军. 山金柑四倍体资源的发掘与鉴定[J]. 园艺学报, 2023, 50(1): 27-35. |
| [3] | 杜玉玲, 杨凡, 赵娟, 刘书琪, 龙超安. 新鱼腥草素钠对柑橘指状青霉的抑菌作用[J]. 园艺学报, 2023, 50(1): 145-152. |
| [4] | 赵雪艳, 王琪, 王莉, 王方圆, 王庆, 李艳. 基于比较转录组的延胡索组织差异性表达分析[J]. 园艺学报, 2023, 50(1): 177-187. |
| [5] | 李镇希, 潘睿翾, 许美容, 郑正, 邓晓玲. 柑橘黄龙病菌双重实时荧光PCR检测方法的建立[J]. 园艺学报, 2023, 50(1): 188-196. |
| [6] | 宋 放, 陈 奇, 袁炎良, 陈 沙, 尹海军, 蒋迎春, . 黄肉猕猴桃新品种‘先沃1号’[J]. 园艺学报, 2022, 49(S2): 47-48. |
| [7] | 齐永杰, 高正辉, 马 娜, 王清明, 柯凡君, 陈 钱, 徐义流, . 黄肉抗溃疡病猕猴桃新品种‘皖农金果’[J]. 园艺学报, 2022, 49(S2): 49-50. |
| [8] | 朱凯杰, 张哲惠, 曹立新, 向舜德, 叶俊丽, 谢宗周, 柴利军, 邓秀新, . 棕色晚熟脐橙新品种‘宗橙’[J]. 园艺学报, 2022, 49(S1): 41-42. |
| [9] | 朱世平, 文荣中, 王媛媛, 曾 杨. 特晚熟柑橘新品种‘金乐柑’[J]. 园艺学报, 2022, 49(S1): 43-44. |
| [10] | 高彦龙, 吴玉霞, 张仲兴, 王双成, 张瑞, 张德, 王延秀. 苹果ELO家族基因鉴定及其在低温胁迫下的表达分析[J]. 园艺学报, 2022, 49(8): 1621-1636. |
| [11] | 邱子文, 刘林敏, 林永盛, 林晓洁, 李永裕, 吴少华, 杨超. 千层金MbEGS基因的克隆与功能分析[J]. 园艺学报, 2022, 49(8): 1747-1760. |
| [12] | 郑林, 王帅, 刘语诺, 杜美霞, 彭爱红, 何永睿, 陈善春, 邹修平. 柑橘响应黄龙病菌侵染的NAC基因的克隆及表达分析[J]. 园艺学报, 2022, 49(7): 1441-1457. |
| [13] | 杨海健, 张云贵, 周心智. 柑橘新品种‘云贵脆橙’[J]. 园艺学报, 2022, 49(7): 1611-1612. |
| [14] | 马维峰, 李艳梅, 马宗桓, 陈佰鸿, 毛娟. 苹果POD家族基因的鉴定与MdPOD15的功能分析[J]. 园艺学报, 2022, 49(6): 1181-1199. |
| [15] | 李文婷, 李翠晓, 林小清, 郑永钦, 郑正, 邓晓玲. 基于STR位点对广东省柑橘溃疡病菌种群遗传结构的分析[J]. 园艺学报, 2022, 49(6): 1233-1246. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司