
园艺学报 ›› 2022, Vol. 49 ›› Issue (5): 995-1007.doi: 10.16420/j.issn.0513-353x.2021-0533
梁晨, 孙如意, 向锐, 孙艺萌, 师校欣, 杜国强( ), 王莉(
), 王莉( )
)
收稿日期:2022-01-27
修回日期:2022-03-28
出版日期:2022-05-25
发布日期:2022-05-25
通讯作者:
杜国强,王莉
E-mail:gdu@hebau.edu.cn;vivi@hebau.edu.cn
基金资助:
LIANG Chen, SUN Ruyi, XIANG Rui, SUN Yimeng, SHI Xiaoxin, DU Guoqiang( ), WANG Li(
), WANG Li( )
)
Received:2022-01-27
Revised:2022-03-28
Online:2022-05-25
Published:2022-05-25
Contact:
DU Guoqiang,WANG Li
E-mail:gdu@hebau.edu.cn;vivi@hebau.edu.cn
摘要:
开展葡萄生长调控因子GRF(Growth-regulating factor)家族成员的基因结构、蛋白保守基序、亚细胞定位预测、同线性、系统进化树及顺式作用元件等分析;利用qRT-PCR技术开展有核和无核葡萄不同组织器官、种子发育不同时期及激素响应表达模式分析。葡萄GRF家族共有8个成员,其中7个分布于6条染色体,编码氨基酸数为213 ~ 604,所有成员均定位于细胞核。根据进化关系分为4组(A ~ D),同组VvGRF的外显子—内含子结构、保守基序数和类型均具有保守性。所有葡萄GRF蛋白N端均含有QLQ和WRC保守结构域,C端至少含有TQL或FFD结构域之一。同线性分析发现,VvGRF3和VvGRF4存在并联重复。VvGRF启动子区发现大量与生长发育、激素响应及胁迫响应相关顺式作用元件。大部分VvGRF在营养器官叶片中高表达,VvGRF2在生殖器官花和果实中高表达;VvGRF3和VvGRF6在无核葡萄种子发育过程中表达量显著高于有核葡萄,而VvGRF8在有核葡萄种子发育前期表达量显著高于无核葡萄;VvGRF响应GA3和IAA诱导,且大部分基因在处理0.5或1 h后下调。
中图分类号:
梁晨, 孙如意, 向锐, 孙艺萌, 师校欣, 杜国强, 王莉. 葡萄生长调控因子GRF家族基因的鉴定及表达分析[J]. 园艺学报, 2022, 49(5): 995-1007.
LIANG Chen, SUN Ruyi, XIANG Rui, SUN Yimeng, SHI Xiaoxin, DU Guoqiang, WANG Li. Genome-wide Identification of Grape GRF Family and Expression Analysis[J]. Acta Horticulturae Sinica, 2022, 49(5): 995-1007.
| 基因Gene | 上游引物(5'-3')Forward primer | 下游引物(5'-3')Reverse primer |
|---|---|---|
| VvGRF1 | GGGCTTCCAGTGCCTTATC | TTCCCATCCGTTCTTCTACAT |
| VvGRF2 | CTGGTGGGACTCATTTCGC | AGGTTGCCTGGCTGTTATTG |
| VvGRF3 | GCCTCAATGACTTCTGAAACCT | CGATGATGCCTGTAATCCCTAT |
| VvGRF4 | CAGAAAGCCTGTGGAAGTTATG | TGTAGCAAGAAGGGTGACGAT |
| VvGRF5 | CGCCTTTGGACAGCACAT | CGAGACTGACCCTCCTGTTTC |
| VvGRF6 | TCGTTCTATCTTCAACCTCCGT | AGTACTTCTGATTGAGTGCCACAT |
| VvGRF7 | GAAGCCCTTGATTCCGTTG | TCTCACAATACTTCTGATCGGC |
| VvGRF8 | CGCCACTCAGGCTATCTCG | AAGCAGGCTCCCATGTTGT |
| EF1-α | AGGAGGCAGCCAACTTCACC | CAAACCCTGCATCACCATTC |
表1 用于qRT-PCR反应的引物序列
Table 1 The primer sequences used for qRT-PCR amplification
| 基因Gene | 上游引物(5'-3')Forward primer | 下游引物(5'-3')Reverse primer |
|---|---|---|
| VvGRF1 | GGGCTTCCAGTGCCTTATC | TTCCCATCCGTTCTTCTACAT |
| VvGRF2 | CTGGTGGGACTCATTTCGC | AGGTTGCCTGGCTGTTATTG |
| VvGRF3 | GCCTCAATGACTTCTGAAACCT | CGATGATGCCTGTAATCCCTAT |
| VvGRF4 | CAGAAAGCCTGTGGAAGTTATG | TGTAGCAAGAAGGGTGACGAT |
| VvGRF5 | CGCCTTTGGACAGCACAT | CGAGACTGACCCTCCTGTTTC |
| VvGRF6 | TCGTTCTATCTTCAACCTCCGT | AGTACTTCTGATTGAGTGCCACAT |
| VvGRF7 | GAAGCCCTTGATTCCGTTG | TCTCACAATACTTCTGATCGGC |
| VvGRF8 | CGCCACTCAGGCTATCTCG | AAGCAGGCTCCCATGTTGT |
| EF1-α | AGGAGGCAGCCAACTTCACC | CAAACCCTGCATCACCATTC |
| 基因 Gene | 登录号 Accession No. | 基因ID Gene locus ID | VCost.v3 ID | 起始与终止位点/bp Start and end position | CDS/bp | ORF/ aa | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|
| VvGRF1 | XP_002281639.1 | GSVIVT01019913001 | Vitvi02g00449 | 4 421 518-4 424 601 | 639 | 213 | 细胞核Nucleus |
| VvGRF2 | XP_002277233.3 | GSVIVT01033800001 | Vitvi08g01498 | 17 725 858-17 728 519 | 1 176 | 392 | 细胞核Nucleus |
| VvGRF3 | XP_010654438.1 | GSVIVT01016762001 | Vitvi09g00107 | 1 135 208-1 137 036 | 1 092 | 364 | 细胞核Nucleus |
| VvGRF4 | XP_010656126.2 | GSVIVT01015095001 | Vitvi11g00092 | 1 000 359-1 001 747 | 1 164 | 388 | 细胞核Nucleus |
| VvGRF5 | XP_002270427.2 | GSVIVT01024326001 | Vitvi16g00073 | 975 755-979 958 | 1 122 | 374 | 细胞核Nucleus |
| VvGRF6 | XP_002271355.3 | GSVIVT01038629001 | Vitvi16g01354 | 21 346 554-21 349 482 | 1 812 | 604 | 细胞核Nucleus |
| VvGRF7 | CBI19365.3 | GSVIVT01009299001 | Vitvi18g00623 | 7 086 633-7 089 964 | 1 428 | 476 | 细胞核Nucleus |
| VvGRF8 | CBI33069.3 | GSVIVT01007165001 | Vitvi07g01551 | 30 596 800-30 600 327 | 1 674 | 558 | 细胞核Nucleus |
表2 葡萄基因组中的GRF家族基因信息
Table 2 Information of GRF family gene in grape genome
| 基因 Gene | 登录号 Accession No. | 基因ID Gene locus ID | VCost.v3 ID | 起始与终止位点/bp Start and end position | CDS/bp | ORF/ aa | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|
| VvGRF1 | XP_002281639.1 | GSVIVT01019913001 | Vitvi02g00449 | 4 421 518-4 424 601 | 639 | 213 | 细胞核Nucleus |
| VvGRF2 | XP_002277233.3 | GSVIVT01033800001 | Vitvi08g01498 | 17 725 858-17 728 519 | 1 176 | 392 | 细胞核Nucleus |
| VvGRF3 | XP_010654438.1 | GSVIVT01016762001 | Vitvi09g00107 | 1 135 208-1 137 036 | 1 092 | 364 | 细胞核Nucleus |
| VvGRF4 | XP_010656126.2 | GSVIVT01015095001 | Vitvi11g00092 | 1 000 359-1 001 747 | 1 164 | 388 | 细胞核Nucleus |
| VvGRF5 | XP_002270427.2 | GSVIVT01024326001 | Vitvi16g00073 | 975 755-979 958 | 1 122 | 374 | 细胞核Nucleus |
| VvGRF6 | XP_002271355.3 | GSVIVT01038629001 | Vitvi16g01354 | 21 346 554-21 349 482 | 1 812 | 604 | 细胞核Nucleus |
| VvGRF7 | CBI19365.3 | GSVIVT01009299001 | Vitvi18g00623 | 7 086 633-7 089 964 | 1 428 | 476 | 细胞核Nucleus |
| VvGRF8 | CBI33069.3 | GSVIVT01007165001 | Vitvi07g01551 | 30 596 800-30 600 327 | 1 674 | 558 | 细胞核Nucleus |
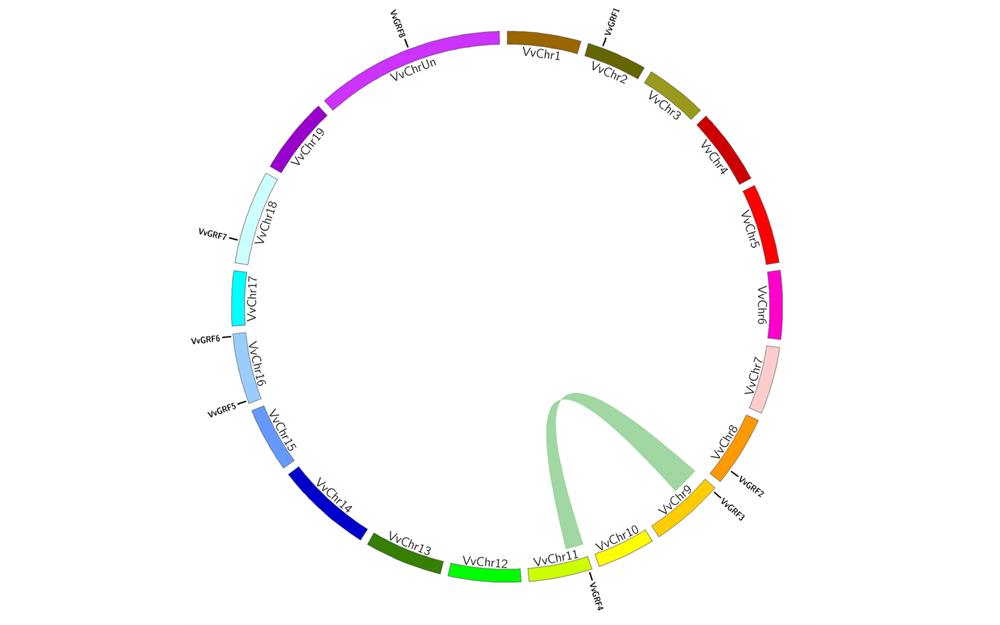
图1 葡萄GRF基因同线性分析 彩带连接的两个染色体区域为同线区域。
Fig. 1 Synteny analysis of grapevine GRF genes Colored lines connection two chromosomal regions are syntenic regions.
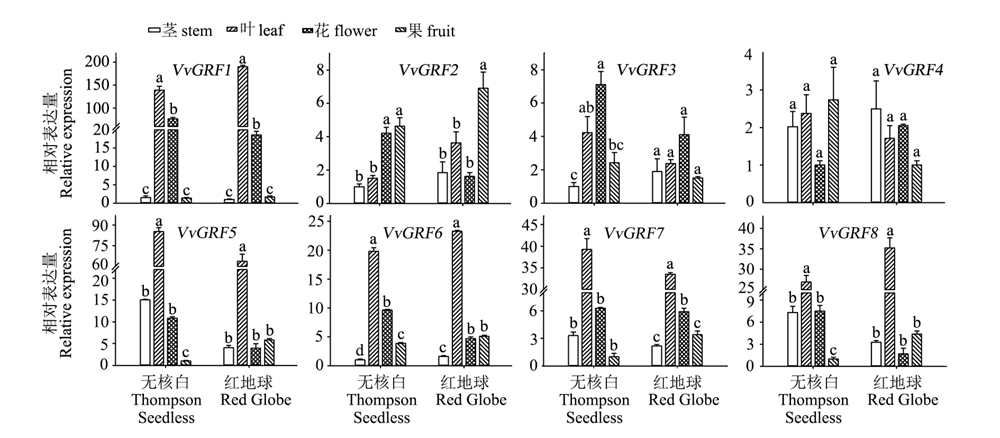
图5 ‘无核白’和‘红地球’葡萄中GRF家族基因在不同器官中的表达(qRT-PCR) 不同字母表示相同品种不同器官间差异显著,Tukey检验方差分析,P < 0.05。
Fig. 5 Expression of GRF family genes in‘Thompson Seedless’and‘Red Globe’grape in different organs(qRT-PCR) Different letters indicate statistically significant differences among different organs in same cultivar. ANOVA with a Tukey post-hoc analysis,P < 0.05.
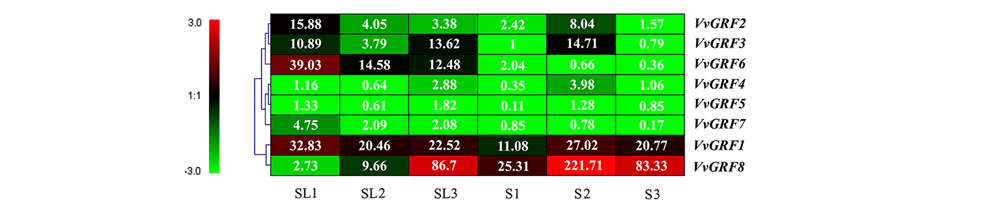
图6 GRF基因在‘红地球’和‘森田尼无核’杂交分离有核(S)和无核(SL)子代种子发育时期的表达
Fig. 6 Expression of GRF genes in the‘Red Globe’and‘Centennial Seedless’hybridization separation seeded(S)and seedless(SL) progenies during seed development

图7 GRF基因在有核和无核葡萄种子发育过程中的表达 不同字母表示相同天数不同品种间差异显著,Tukey检验方差分析,P < 0.05。
Fig. 7 Expression of GRF genes in seeded and seedless grapes during seed development Different letters indicate significant differences among different cultivars in the same day. ANOVA with a Tukey post-hoc analysis,P < 0.05.
| [1] |
Ai G, Zhang D, Huang R, Zhang S, Li W, Ahiakpa J K, Zhang J. 2020. Genome-wide identification and molecular characterization of the growth-regulating factors-interacting factor gene family in tomato. Genes, 11 (12):1435.
doi: 10.3390/genes11121435 URL |
| [2] | Bailey T L, Boden M, Buske F A, Frith M, Grant C E, Clementi L, Ren J, Li W W, Noble W S. 2009. MEME SUITE:tools for motif discovery and searching. Nucleic Acids Research, 37 (s2):W202-W208. |
| [3] | Cao Y, Han Y, Jin Q, Lin Y, Cai Y. 2016. Comparative genomic analysis of the GRF genes in Chinese pear(Pyrus bretschneideri Rehd),poplar(Populous),grape(Vitis vinifera),Arabidopsis and rice(Oryza sativa). Frontiers in Plant Science, 7:1750. |
| [4] |
Chen C, Chen H, Zhang Y, Thomas H R, Frank M H, He Y, Xia R. 2020. TBtools:an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 13 (8):1194-1202.
doi: 10.1016/j.molp.2020.06.009 URL |
| [5] |
Choi D, Kim J H, Kende H. 2004. Whole genome analysis of the OsGRF gene family encoding plant-specific putative transcription activators in rice(Oryza sativa L.). Plant and Cell Physiology, 45 (7):897-904.
doi: 10.1093/pcp/pch098 URL |
| [6] |
Horiguchi G, Kim G T, Tsukaya H. 2005. The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant Journal, 43 (1):68-78.
pmid: 15960617 |
| [7] |
Howe E A, Sinha R, Schlauch D, Quackenbush J. 2011. RNA-Seq analysis in MeV. Bioinformatics, 27 (22):3209-3210.
doi: 10.1093/bioinformatics/btr490 URL |
| [8] |
Hu B, Jin J, Guo A Y, Zhang H, Luo J, Gao G. 2015. GSDS 2.0:an upgraded gene feature visualization server. Bioinformatics, 31 (8):1296-1297.
doi: 10.1093/bioinformatics/btu817 URL |
| [9] |
Khatun K, Robin A H K, Park J I, Nath U K, Kim C K, Lim K B, Nou I S, Chung M Y. 2017. Molecular characterization and expression profiling of tomato GRF transcription factor family genes in response to abiotic stresses and phytohormones. International Journal of Molecular Sciences, 18 (5):1056.
doi: 10.3390/ijms18051056 URL |
| [10] |
Kim J H, Choi D, Kende H. 2003. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant Journal, 36 (1):94-104.
doi: 10.1046/j.1365-313X.2003.01862.x URL |
| [11] | Kim J H, Kende H. 2004. A transcriptional coactivator,AtGIF1,is involved in regulating leaf growth and morphology in Arabidopsis. Proceedings of the National Academy of Sciences, 101 (36):13374-13379. |
| [12] |
Kim J H, Lee B H. 2006. Growth-regulating factor 4 of Arabidopsis thaliana is required for development of leaves,cotyledons,and shoot apical meristem. Journal of Plant Biology, 49 (6):463-468.
doi: 10.1007/BF03031127 URL |
| [13] |
Kim J S, Mizoi J, Kidokoro S, Maruyama K, Nakajima J, Nakashima K, Mitsuda N, Takiguchi Y, Ohme-Takagi M, Kondou Y, Yoshizumi T, Matsui M, Shinozaki K, Yamaguchi-Shinozaki K. 2012. Arabidopsis GROWTH-REGULATING FACTOR7 functions as a transcriptional repressor of abscisic acid- and osmotic stress-responsive genes,including DREB2A. Plant Cell, 24 (8):3393-3405.
doi: 10.1105/tpc.112.100933 URL |
| [14] |
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones S J, Marra M A. 2009. Circos:an information aesthetic for comparative genomics. Genome Research, 19 (9):1639-1645.
doi: 10.1101/gr.092759.109 pmid: 19541911 |
| [15] |
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X:molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35 (6):1547-1549.
doi: 10.1093/molbev/msy096 URL |
| [16] | Lee T H, Tang H, Wang X, Paterson A H. 2013. PGDD:a database of gene and genome duplication in plants. Nucleic Acids Research, 41:D1152-1158. |
| [17] | Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. 2002. PlantCARE,a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research, 30 (1):325-327. |
| [18] |
Li S, Gao F, Xie K, Zeng X, Cao Y, Zeng J, He Z, Ren Y, Li W, Deng Q, Wang S, Zheng A, Zhu J, Liu H, Wang L, Li P. 2016. The OsmiR396c-OsGRF4-OsGIF 1 regulatory module determines grain size and yield in rice. Plant Biotechnology Journal, 14 (11):2134-2146.
doi: 10.1111/pbi.12569 URL |
| [19] | Liu Pudong, Zhang Shuting, Chen Xiaohui, Su Liyao, Yao Deheng, Li Hansheng, Zhang Zihao. 2020. Genomic identification and expression patterns of the longan GRF family. Chinese Journal of Applied and Environmental Biology, 26 (2):236-245. (in Chinese) |
| 刘蒲东, 张舒婷, 陈晓慧, 苏立遥, 姚德恒, 李汉生, 张梓浩, 陈裕坤, 林玉玲, 赖钟雄. 2020. 龙眼GRF家族全基因组鉴定及表达模式. 应用与环境生物学报, 26 (2):236-245. | |
| [20] |
Liu X, Guo L X, Jin L F, Liu Y Z, Liu T, Fan Y H, Peng S A. 2016. Identification and transcript profiles of citrus growth-regulating factor genes involved in the regulation of leaf and fruit development. Molecular Biology Reports, 43 (10):1059-1067.
doi: 10.1007/s11033-016-4048-1 URL |
| [21] |
Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods, 25 (4):402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [22] |
Omidbakhshfard M A, Proost S, Fujikura U, Mueller-Roeber B. 2015. Growth-regulating factors(GRFs):a small transcription factor family with important functions in plant biology. Molecular Plant, 8 (7):998-1010.
doi: 10.1016/j.molp.2015.01.013 pmid: 25620770 |
| [23] |
Sun P, Zhang W, Wang Y, He Q, Shu F, Liu H, Wang J, Wang J, Yuan L, Deng H. 2016. OsGRF4 controls grain shape,panicle length and seed shattering in rice. Journal of Integrative Plant Biology, 58 (10):836-847.
doi: 10.1111/jipb.12473 URL |
| [24] |
van der Knaap E, Kim J H, Kende H. 2000. A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiology, 122 (3):695-704.
pmid: 10712532 |
| [25] |
Wang F, Qiu N, Ding Q, Li J, Zhang Y, Li H, Gao J. 2014. Genome-wide identification and analysis of the growth-regulating factor family in Chinese cabbage(Brassica rapa L. ssp. pekinensis). BMC Genomics, 15:807.
doi: 10.1186/1471-2164-15-807 URL |
| [26] |
Wang L, Ahmad B, Liang C, Shi X X, Sun R Y, Zhang S L, Du G Q. 2020a. Bioinformatics and expression analysis of histone modification genes in grapevine predict their involvement in seed development,powdery mildew resistance,and hormonal signaling. BMC Plant Biology, 20 (1):412.
doi: 10.1186/s12870-020-02618-7 URL |
| [27] |
Wang L, Hu X, Jiao C, Li Z, Fei Z, Yan X, Liu C, Wang Y, Wang X. 2016. Transcriptome analyses of seed development in grape hybrids reveals a possible mechanism influencing seed size. BMC Genomics, 17 (1):898.
pmid: 27829355 |
| [28] |
Wang L, Yin X, Cheng C, Wang H, Guo R, Xu X, Zhao J, Zheng Y, Wang X. 2015. Evolutionary and expression analysis of a MADS-box gene superfamily involved in ovule development of seeded and seedless grapevines. Molecular Genetics and Genomics, 290 (3):825-846.
doi: 10.1007/s00438-014-0961-y pmid: 25429734 |
| [29] |
Wang L, Zhang S L, Zhang X M, Hu X Y, Guo C L, Wang X P, Song J Y. 2018. Evolutionary and expression analysis of Vitis vinifera OFP gene family. Plant Systematics and Evolution, 304 (8):995-1008.
doi: 10.1007/s00606-018-1528-x URL |
| [30] |
Wang J N, Zhou H J, Zhao Y Q, Sun P B, Tang F, Song X Q, Lu M Z. 2020b. Characterization of poplar growth regulating factors and analysis of their function in leaf size control. BMC Plant Biology, 20:509.
doi: 10.1186/s12870-020-02699-4 URL |
| [31] | Wang Xiaofang, Chang Qinxiang, Wang Shufang, Yan Zhao, Li Xuyan, Gary Gao, Ji Wei. 2021. Bioinformatics analysis of GRF gene family in grape. Molecular Plant Breeding, 19 (18):5975-5983. (in Chinese) |
| 王晓芳, 昌秦湘, 王淑芳, 闫钊, 李戌彦, Gary Gao, 纪薇. 2021. 葡萄GRF基因家族的生物信息学分析. 分子植物育种, 19 (18):5975-5983. | |
| [32] | Wei Yanhong, Liu Zhen, Li Ke, Meng Yuan, Wang Hui, Mao Jiangping, Ma Doudou, Li Shaohuan, Ma Juanjuan, Lu Xian, Zhang Dong. 2020. Genome-wide identification and expression analysis of miR396 family during adventitious root development in apple. Acta Horticulturae Sinica, 47 (7):1237-1252. (in Chinese) |
| 韦燕红, 刘桢, 李珂, 孟媛, 汪蕙, 毛江萍, 马豆豆, 李少欢, 马娟娟, 卢显, 张东. 2020. 苹果miR396 家族鉴定及在不定根发育过程中的表达分析. 园艺学报, 47 (7):1237-1252. | |
| [33] |
Wu L, Zhang D, Xue M, Qian J, He Y, Wang S. 2014. Overexpression of the maize GRF10,an endogenous truncated growth-regulating factor protein,leads to reduction in leaf size and plant height. Journal of Integrative Plant Biology, 56 (11):1053-1063.
doi: 10.1111/jipb.12220 URL |
| [34] | Wu Z J, Wang W L, Zhuang J. 2017. Developmental processes and responses to hormonal stimuli in tea plant(Camellia sinensis)leaves are controlled by GRF and GIF gene families. Functional & Integrative Genomics, 17 (5):503-512. |
| [35] |
Zhang D F, Li B, Jia G Q, Zhang T F, Dai J R, Li J S, Wang S C. 2008. Isolation and characterization of genes encoding GRF transcription factors and GIF transcriptional coactivators in maize(Zea mays L.). Plant Science, 175 (6):809-817.
doi: 10.1016/j.plantsci.2008.08.002 URL |
| [36] |
Zhang J, Li Z, Jin J, Xie X, Zhang H, Chen Q, Luo Z, Yang J. 2018. Genome-wide identification and analysis of the growth-regulating factor family in tobacco(Nicotiana tabacum). Gene, 639:117-127.
doi: 10.1016/j.gene.2017.09.070 URL |
| [37] |
Zhang S, Wang L, Sun X, Li Y, Yao J, van Nocker S, Wang X. 2019. Genome-wide analysis of the YABBY gene family in grapevine and functional characterization of VvYABBY4. Frontiers in Plant Science, 10:1207.
doi: 10.3389/fpls.2019.01207 URL |
| [38] | Zhao Huixia, Li Fengxia, Guo Rui, Chen Chanyou. 2020. Genome-wide characterization and expression analysis of GRF gene family in Capsicum. Acta Horticulturae Sinica, 47 (11):2145-2160. (in Chinese) |
| 赵慧霞, 李凤霞, 郭瑞, 陈禅友. 2020. 辣椒属GRF基因家族全基因组鉴定和表达分析. 园艺学报, 47 (11):2145-2160. | |
| [39] | Zheng L W, Ma J J, Song C H, Zhang L Z, Gao C, Zhang D, An N, Mao J P, Han M Y. 2018. Genome-wide identification and expression analysis of GRF genes regulating apple tree architecture. Tree Genetics & Genomes, 14 (4):54. |
| [40] |
Zhou H J, Song X Q, Wei K L, Zhao Y Q, Jiang C, Wang J N, Tang F, Lu M Z. 2019. Growth-regulating factor 15 is required for leaf size control in Populus. Tree Physiology, 39 (3):381-390.
doi: 10.1093/treephys/tpy107 URL |
| [41] |
Zhu Y X, Li Y D, Zhang S L, Zhang X M, Yao J, Luo Q W, Sun F, Wang X P. 2019. Genome-wide identification and expression analysis reveal the potential function of ethylene responsive factor gene family in response to Botrytis cinerea infection and ovule development in grapes(Vitis vinifera L.). Plant Biology, 21 (4):571-584.
doi: 10.1111/plb.12943 pmid: 30468551 |
| [1] | 王晓晨, 聂子页, 刘先菊, 段 伟, 范培格, 梁振昌, . 脱落酸对‘京香玉’葡萄果实单萜物质合成的影响[J]. 园艺学报, 2023, 50(2): 237-249. |
| [2] | 袁馨, 徐云鹤, 张雨培, 单楠, 陈楚英, 万春鹏, 开文斌, 翟夏琬, 陈金印, 甘增宇. 猕猴桃后熟过程中ABA响应结合因子AcAREB1调控AcGH3.1的表达[J]. 园艺学报, 2023, 50(1): 53-64. |
| [3] | 赵雪艳, 王琪, 王莉, 王方圆, 王庆, 李艳. 基于比较转录组的延胡索组织差异性表达分析[J]. 园艺学报, 2023, 50(1): 177-187. |
| [4] | 王宝亮, 刘凤之, 冀晓昊, 王孝娣, 史祥宾, 张艺灿, 李 鹏, 王海波. 早熟鲜食葡萄新品种‘华葡早玉’[J]. 园艺学报, 2022, 49(S2): 33-34. |
| [5] | 王宝亮, 王海波, 冀晓昊, 王孝娣, 史祥宾, 王志强, 王小龙, 刘凤之. 中熟鲜食葡萄新品种‘华葡黄玉’[J]. 园艺学报, 2022, 49(S2): 35-36. |
| [6] | 牛早柱, 赵艳卓, 陈 展, 宣立锋, 牛帅科, 魏建国, 褚凤杰, 杨丽丽. 晚熟无核葡萄新品种‘紫龙珠’[J]. 园艺学报, 2022, 49(S2): 37-38. |
| [7] | 师校欣, 杜国强, 杨丽丽, 乔月莲, 黄成立, 王素月, 赵跃欣, 魏晓慧, 王 莉, 齐向丽. 晚熟无核葡萄新品种‘红峰无核’[J]. 园艺学报, 2022, 49(S2): 39-40. |
| [8] | 吴月燕, 陈天池, 王立如, 韩善琪, 付 涛. 鲜食葡萄新品种‘甬早红’[J]. 园艺学报, 2022, 49(S2): 41-42. |
| [9] | 王晓玥, 闫爱玲, 张国军, 王慧玲, 任建成, 刘振华, 孙 磊, 徐海英, . 葡萄新品种‘瑞都晚红’[J]. 园艺学报, 2022, 49(S1): 29-30. |
| [10] | 高彦龙, 吴玉霞, 张仲兴, 王双成, 张瑞, 张德, 王延秀. 苹果ELO家族基因鉴定及其在低温胁迫下的表达分析[J]. 园艺学报, 2022, 49(8): 1621-1636. |
| [11] | 邱子文, 刘林敏, 林永盛, 林晓洁, 李永裕, 吴少华, 杨超. 千层金MbEGS基因的克隆与功能分析[J]. 园艺学报, 2022, 49(8): 1747-1760. |
| [12] | 王勇健, 孔俊花, 范培格, 梁振昌, 金秀良, 刘布春, 代占武. 葡萄表型组高通量获取及分析方法研究进展[J]. 园艺学报, 2022, 49(8): 1815-1832. |
| [13] | 郑林, 王帅, 刘语诺, 杜美霞, 彭爱红, 何永睿, 陈善春, 邹修平. 柑橘响应黄龙病菌侵染的NAC基因的克隆及表达分析[J]. 园艺学报, 2022, 49(7): 1441-1457. |
| [14] | 马维峰, 李艳梅, 马宗桓, 陈佰鸿, 毛娟. 苹果POD家族基因的鉴定与MdPOD15的功能分析[J]. 园艺学报, 2022, 49(6): 1181-1199. |
| [15] | 魏晓羽, 王跃进. 中国野生葡萄果皮解剖结构与白粉病抗性的相关性研究[J]. 园艺学报, 2022, 49(6): 1200-1212. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司