
园艺学报 ›› 2022, Vol. 49 ›› Issue (4): 827-840.doi: 10.16420/j.issn.0513-353x.2020-1069
王莹, 艾鹏慧, 李帅磊, 康冬茹, 李忠爱, 王子成**( )
)
收稿日期:2021-07-16
修回日期:2021-11-15
出版日期:2022-04-25
发布日期:2022-04-24
通讯作者:
王子成
E-mail:wzc@henu.edu.cn
基金资助:
WANG Ying, AI Penghui, LI Shuailei, KANG Dongru, LI Zhongai, WANG Zicheng**( )
)
Received:2021-07-16
Revised:2021-11-15
Online:2022-04-25
Published:2022-04-24
Contact:
WANG Zicheng
E-mail:wzc@henu.edu.cn
摘要:
基于传统菊花(Chrysanthemum × morifolium Ramat.)品种‘金背大红’转录组数据和菊花脑(C. nankingense)的基因组数据,通过生物信息学方法鉴定了二者的DNA甲基化相关酶基因,分析其编码蛋白结构域并构建系统发育树,还分析了基因的表达情况。从‘金背大红’中鉴定出了13个DNA甲基转移酶基因和11个去甲基化酶基因,从菊花脑中鉴定出10个DNA甲基转移酶基因和5个去甲基化酶基因。MET1同源氨基酸序列系统进化分析结果表明,菊花‘紫精灵’‘金背大红’和‘禾城星火’与甘菊[C. lavandulifolium(Fisch. ex Trautv.)Ling et Shih]处于较近的分支中,这表明他们可能有更近的演化关系,而菊花脑和菊花‘泉乡水长’与甘菊的进化关系可能较远。DNA甲基化相关酶基因在不同组织及不同菊花品种中的表达水平存在较大差异。
中图分类号:
王莹, 艾鹏慧, 李帅磊, 康冬茹, 李忠爱, 王子成. 菊花和菊花脑DNA甲基化相关酶基因鉴定及表达分析[J]. 园艺学报, 2022, 49(4): 827-840.
WANG Ying, AI Penghui, LI Shuailei, KANG Dongru, LI Zhongai, WANG Zicheng. Identification and Expression Analysis of Genes Related to DNA Methylation in Chrysanthemum × morifolium and C. nankingense[J]. Acta Horticulturae Sinica, 2022, 49(4): 827-840.
| 基因 Gene | 上游引物序列(5′-3′) Forward primer sequence | 下游引物序列(5′-3′) Reverse primer sequence |
|---|---|---|
| Cm-methylase1 | AACTGGTCACTATACAACTG | ATCATCTGCCGTCATTAC |
| Cm-methylase2 | GACGCTCTCCAAGATTAG | ACCACCATCATTAGATTCAA |
| Cm-methylase10 | TGACTCCGATAGGCTACTT | GGTATCCGAATCAATATCAACATT |
| Cm-demethylase3 | TGGAGAATGATGGAAGTG | GCCTGTTACCTTGGATAA |
| Cn-methylase1 | TGTCAAAATCTGCTGGAGG | TTGTCATTATTGTTGGGGG |
| Cn-methylase2 | AAAAAAGAAACGAAAGGGTC | ATAAAGAAAGCGAGAAATGG |
| Cn-methylase3 | TACAATTCATGGTGCCTTGC | ATTTTGCGCTTCCATATGCT |
| Cn-methylase4 | GCTTCTTCCACTTCCTCCCC | CTTAAACCCCCACCTCTCGT |
| Cn-methylase5 | TTGCCTCAGTTTCCTTTACC | AGCACCTCCATACTCCATTT |
| Cn-methylase6 | AATCACTTTGCTATCTCTCT | TTACTTACTTGTGTCTCTGG |
| Cn-methylase7 | TCTTTTGCTCTTGCTCACAC | CACACACAGCCCACACATTT |
| Cn-methylase8 | CCAAGCTAATGTGTGGCTTC | GCTGTCCATTAAACTTTGGG |
| Cn-methylase9 | TTTTGGAAGTGAATACAGGG | ATGGTTGACATAGTGGAGAA |
| Cn-methylase10 | ATGACAGTGAAGGACAGCGA | GAAATGAGGGGTATGAAAGG |
| Cn-demethylase1 | CAAACCTAAGAAACCGCCAA | CAACATCTCCAAAGCCACAA |
| Cn-demethylase2 | CTGAAGAAACTGGTAGACAA | CAAAATCAAGAACCTAATGG |
| Cn-demethylase3 | ACAGATAAGGACCACGAATG | GGACTGAATAACCAGCAAAC |
| Cn-demethylase4 | CCACCAGAAGGAAAATGTCG | CTCACCAGGATTGTCCCACG |
| Cn-demethylase5 | AAACCGAAGCCAATACTGAG | TTCTAAGAGTGGGTGGGAGT |
| Actin | AGCTTGCATATGTTGCTCTTGA | TTACCGTAAAGGTCCTTCCTGA |
表1 荧光定量引物序列
Table 1 Appendix a sequences for fluorescent quantitative
| 基因 Gene | 上游引物序列(5′-3′) Forward primer sequence | 下游引物序列(5′-3′) Reverse primer sequence |
|---|---|---|
| Cm-methylase1 | AACTGGTCACTATACAACTG | ATCATCTGCCGTCATTAC |
| Cm-methylase2 | GACGCTCTCCAAGATTAG | ACCACCATCATTAGATTCAA |
| Cm-methylase10 | TGACTCCGATAGGCTACTT | GGTATCCGAATCAATATCAACATT |
| Cm-demethylase3 | TGGAGAATGATGGAAGTG | GCCTGTTACCTTGGATAA |
| Cn-methylase1 | TGTCAAAATCTGCTGGAGG | TTGTCATTATTGTTGGGGG |
| Cn-methylase2 | AAAAAAGAAACGAAAGGGTC | ATAAAGAAAGCGAGAAATGG |
| Cn-methylase3 | TACAATTCATGGTGCCTTGC | ATTTTGCGCTTCCATATGCT |
| Cn-methylase4 | GCTTCTTCCACTTCCTCCCC | CTTAAACCCCCACCTCTCGT |
| Cn-methylase5 | TTGCCTCAGTTTCCTTTACC | AGCACCTCCATACTCCATTT |
| Cn-methylase6 | AATCACTTTGCTATCTCTCT | TTACTTACTTGTGTCTCTGG |
| Cn-methylase7 | TCTTTTGCTCTTGCTCACAC | CACACACAGCCCACACATTT |
| Cn-methylase8 | CCAAGCTAATGTGTGGCTTC | GCTGTCCATTAAACTTTGGG |
| Cn-methylase9 | TTTTGGAAGTGAATACAGGG | ATGGTTGACATAGTGGAGAA |
| Cn-methylase10 | ATGACAGTGAAGGACAGCGA | GAAATGAGGGGTATGAAAGG |
| Cn-demethylase1 | CAAACCTAAGAAACCGCCAA | CAACATCTCCAAAGCCACAA |
| Cn-demethylase2 | CTGAAGAAACTGGTAGACAA | CAAAATCAAGAACCTAATGG |
| Cn-demethylase3 | ACAGATAAGGACCACGAATG | GGACTGAATAACCAGCAAAC |
| Cn-demethylase4 | CCACCAGAAGGAAAATGTCG | CTCACCAGGATTGTCCCACG |
| Cn-demethylase5 | AAACCGAAGCCAATACTGAG | TTCTAAGAGTGGGTGGGAGT |
| Actin | AGCTTGCATATGTTGCTCTTGA | TTACCGTAAAGGTCCTTCCTGA |
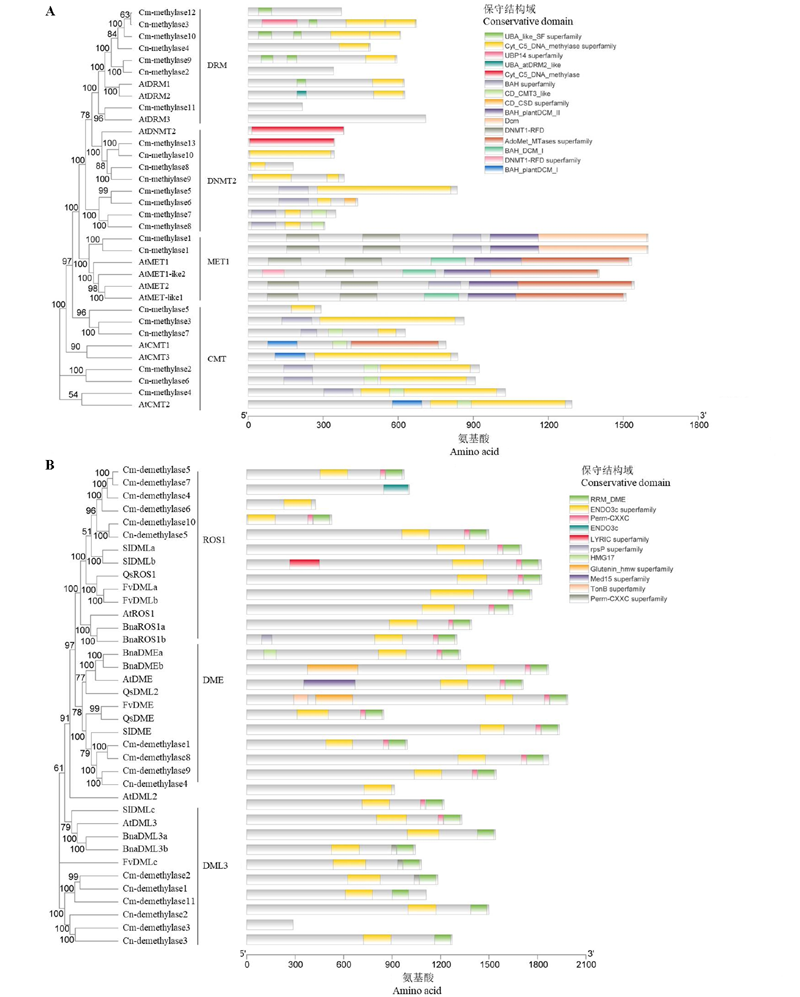
图1 ‘金背大红’菊(Cm)和菊花脑(Cn)DNA甲基转移酶(A)和去甲基化酶(B)的系统发育树和保守结构域 采用MEGA7,邻接聚类方法,Bootstrap的值为1 000。数字表示自展值。 At:拟南芥;Sl:番茄;Qs:欧洲栓皮栎;Fv:森林草莓;Bna:甘蓝型油菜。
Fig. 1 Phylogenetic tree and conserved domains of DNA methylases(A)and demethylases(B)of Chrysanthemum ‘Jinbeidahong’(Cm)and Chrysanthemum nankingense(Cn) Using the Neighbour-joining method in MEGA 7 software. The value of bootstrap is 1 000. Numbers indicate bootstrap values. At:Arabidopsis thaliana;Sl:Solanum lycopersicum;Qs:Quercus suber;Fv:Fragaria vesca;Bna:Brassica napus.
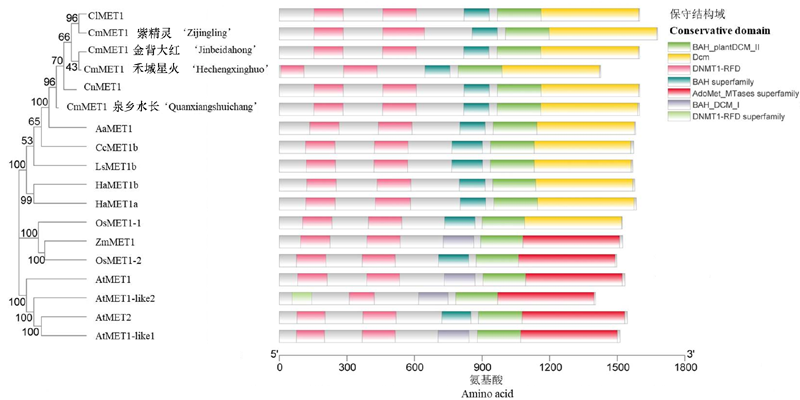
图2 不同物种中MET1的系统发育树和保守结构域 采用MEGA7,邻接聚类方法,Bootstrap的值为1 000。数字表示自展值。 Cl:甘菊;Cn:菊花脑;Aa:青蒿;Cc:刺苞菜蓟;Ls:莴苣;Ha:向日葵;Os:水稻;Zm:玉米;At:拟南芥。
Fig. 2 Phylogenetic tree and conserved domains of MET1 family in different species Using the Neighbour-joining method in MEGA 7 software. The value of bootstrap is 1 000. Numbers indicate bootstrap values. Cl:C. lavandulifolium;Cn:C. nankingense;Aa:Artemisia annua;Cc:Cynara cardunculus;Ls:Lactuca saliva;Ha:Helianthus annuus;Os:Oryza sativa;Zm:Zea mays;At:Arabidopsis thaliana.
| 酶 Enzyme | 基因 Gene name | CDS长度/bp CDS length | 蛋白质 Protein | 类型 Type | ||
|---|---|---|---|---|---|---|
| 长度/aa Length | 分子量/Da Mw | 等电点 pI | ||||
| 甲基转移酶 | Cm-methylase1 | 4 800 | 1 599 | 178 739.29 | 5.75 | MET1 |
| Methyltransferase | Cm-methylase2 | 2 788 | 925 | 103 843.57 | 6.14 | CMT |
| Cm-methylase3 | 2 595 | 864 | 97 299.75 | 4.91 | CMT | |
| Cm-methylase4 | 3 093 | 1 030 | 116 027.35 | 5.92 | CMT | |
| Cm-methylase5 | 2 511 | 836 | 94 321.79 | 5.95 | DNMT2 | |
| Cm-methylase6 | 1 317 | 438 | 49 870.56 | 5.24 | DNMT2 | |
| Cm-methylase7 | 1 053 | 350 | 39 764.17 | 5.13 | DNMT2 | |
| Cm-methylase8 | 921 | 306 | 34 963.24 | 4.91 | DNMT2 | |
| Cm-methylase9 | 1 788 | 595 | 66 491.85 | 4.89 | DRM | |
| Cm-methylase10 | 1 833 | 610 | 68 071.51 | 4.96 | DRM | |
| Cm-methylase11 | 654 | 217 | 24 671.57 | 8.97 | DRM | |
| Cm-methylase12 | 1 122 | 373 | 40 971.16 | 4.34 | DRM | |
| Cm-methylase13 | 1 041 | 346 | 39 705.42 | 5.83 | DNMT2 | |
| 去甲基化酶 | Cm-demethylase1 | 4 638 | 1 545 | 173 065.09 | 8.26 | DME |
| Demethylase | Cm-demethylase2 | 4 503 | 1 500 | 170 278.85 | 6.35 | DML3 |
| Cm-demethylase3 | 3 822 | 1 273 | 144 744.28 | 8.93 | DML3 | |
| Cm-demethylase4 | 1 281 | 426 | 47 988.14 | 5.92 | ROS1 | |
| Cm-demethylase5 | 2 928 | 975 | 110 623.30 | 5.53 | ROS1 | |
| Cm-demethylase6 | 1 584 | 527 | 60 170.57 | 5.78 | ROS1 | |
| Cm-demethylase7 | 3 027 | 1 008 | 115 750.42 | 5.78 | ROS1 | |
| Cm-demethylase8 | 2 751 | 916 | 103 280.83 | 8.51 | DME | |
| Cm-demethylase9 | 3 672 | 1 223 | 137 284.38 | 6.74 | DME | |
| Cm-demethylase10 | 4 503 | 1 500 | 169 026.50 | 6.05 | ROS1 | |
| Cm-demethylase11 | 864 | 287 | 32 569.63 | 4.37 | DML3 | |
表2 ‘金背大红’菊的DNA甲基转移酶基因和去甲基化酶基因的结构分析
Table 2 Structural analysis of DNA methyltransferases and demethylase genes of Chrysanthemum‘Jinbeidahong’
| 酶 Enzyme | 基因 Gene name | CDS长度/bp CDS length | 蛋白质 Protein | 类型 Type | ||
|---|---|---|---|---|---|---|
| 长度/aa Length | 分子量/Da Mw | 等电点 pI | ||||
| 甲基转移酶 | Cm-methylase1 | 4 800 | 1 599 | 178 739.29 | 5.75 | MET1 |
| Methyltransferase | Cm-methylase2 | 2 788 | 925 | 103 843.57 | 6.14 | CMT |
| Cm-methylase3 | 2 595 | 864 | 97 299.75 | 4.91 | CMT | |
| Cm-methylase4 | 3 093 | 1 030 | 116 027.35 | 5.92 | CMT | |
| Cm-methylase5 | 2 511 | 836 | 94 321.79 | 5.95 | DNMT2 | |
| Cm-methylase6 | 1 317 | 438 | 49 870.56 | 5.24 | DNMT2 | |
| Cm-methylase7 | 1 053 | 350 | 39 764.17 | 5.13 | DNMT2 | |
| Cm-methylase8 | 921 | 306 | 34 963.24 | 4.91 | DNMT2 | |
| Cm-methylase9 | 1 788 | 595 | 66 491.85 | 4.89 | DRM | |
| Cm-methylase10 | 1 833 | 610 | 68 071.51 | 4.96 | DRM | |
| Cm-methylase11 | 654 | 217 | 24 671.57 | 8.97 | DRM | |
| Cm-methylase12 | 1 122 | 373 | 40 971.16 | 4.34 | DRM | |
| Cm-methylase13 | 1 041 | 346 | 39 705.42 | 5.83 | DNMT2 | |
| 去甲基化酶 | Cm-demethylase1 | 4 638 | 1 545 | 173 065.09 | 8.26 | DME |
| Demethylase | Cm-demethylase2 | 4 503 | 1 500 | 170 278.85 | 6.35 | DML3 |
| Cm-demethylase3 | 3 822 | 1 273 | 144 744.28 | 8.93 | DML3 | |
| Cm-demethylase4 | 1 281 | 426 | 47 988.14 | 5.92 | ROS1 | |
| Cm-demethylase5 | 2 928 | 975 | 110 623.30 | 5.53 | ROS1 | |
| Cm-demethylase6 | 1 584 | 527 | 60 170.57 | 5.78 | ROS1 | |
| Cm-demethylase7 | 3 027 | 1 008 | 115 750.42 | 5.78 | ROS1 | |
| Cm-demethylase8 | 2 751 | 916 | 103 280.83 | 8.51 | DME | |
| Cm-demethylase9 | 3 672 | 1 223 | 137 284.38 | 6.74 | DME | |
| Cm-demethylase10 | 4 503 | 1 500 | 169 026.50 | 6.05 | ROS1 | |
| Cm-demethylase11 | 864 | 287 | 32 569.63 | 4.37 | DML3 | |
| 酶 Enzyme | 基因 Gene Name | CDS长度/bp CDS length | 外显子数 Number of exons | 蛋白质Protein | 类型 Type | ||
|---|---|---|---|---|---|---|---|
| 长度/aa Length | 分子量/Da Mw | 等电点 pI | |||||
| 甲基转移酶 | Cn-methylase1 | 4 803 | 3 | 1 600 | 178 962.60 | 5.78 | MET1 |
| Methyltransferase | Cn-methylase2 | 1 029 | 4 | 342 | 39 472.94 | 9.68 | DRM |
| Cn-methylase3 | 2 019 | 12 | 672 | 75 038.30 | 5.01 | DRM | |
| Cn-methylase4 | 1 470 | 9 | 489 | 54 992.14 | 5.19 | DRM | |
| Cn-methylase5 | 879 | 10 | 292 | 32 439.25 | 7.63 | CMT | |
| Cn-methylase6 | 2 730 | 20 | 909 | 101 888.25 | 6.20 | CMT | |
| Cn-methylase7 | 1 887 | 18 | 628 | 71 372.50 | 4.99 | CMT | |
| Cn-methylase8 | 543 | 4 | 181 | 20 947.95 | 7.57 | DNMT2 | |
| Cn-methylase9 | 1 155 | 9 | 384 | 44 105.80 | 6.36 | DNMT2 | |
| Cn-methylase10 | 1 038 | 9 | 345 | 39 700.18 | 5.27 | DNMT2 | |
| 去甲基化酶 | Cn-demethylase1 | 4 254 | 18 | 1 417 | 160 835.45 | 7.46 | DML3 |
| Demethylase | Cn-demethylase2 | 2 805 | 14 | 934 | 106 836.11 | 8.79 | DML3 |
| Cn-demethylase3 | 2 613 | 15 | 870 | 99 036.82 | 9.20 | DML3 | |
| Cn-demethylase4 | 5 244 | 20 | 1 747 | 194 573.45 | 6.65 | DME | |
| Cn-demethylase5 | 4 707 | 18 | 1 568 | 175 882.89 | 5.96 | ROS1 | |
表3 菊花脑的DNA甲基转移酶基因和去甲基化酶基因的结构分析
Table 3 Structural analysis of DNA methyltransferases and demethylase genes of Chrysanthemum nankingense
| 酶 Enzyme | 基因 Gene Name | CDS长度/bp CDS length | 外显子数 Number of exons | 蛋白质Protein | 类型 Type | ||
|---|---|---|---|---|---|---|---|
| 长度/aa Length | 分子量/Da Mw | 等电点 pI | |||||
| 甲基转移酶 | Cn-methylase1 | 4 803 | 3 | 1 600 | 178 962.60 | 5.78 | MET1 |
| Methyltransferase | Cn-methylase2 | 1 029 | 4 | 342 | 39 472.94 | 9.68 | DRM |
| Cn-methylase3 | 2 019 | 12 | 672 | 75 038.30 | 5.01 | DRM | |
| Cn-methylase4 | 1 470 | 9 | 489 | 54 992.14 | 5.19 | DRM | |
| Cn-methylase5 | 879 | 10 | 292 | 32 439.25 | 7.63 | CMT | |
| Cn-methylase6 | 2 730 | 20 | 909 | 101 888.25 | 6.20 | CMT | |
| Cn-methylase7 | 1 887 | 18 | 628 | 71 372.50 | 4.99 | CMT | |
| Cn-methylase8 | 543 | 4 | 181 | 20 947.95 | 7.57 | DNMT2 | |
| Cn-methylase9 | 1 155 | 9 | 384 | 44 105.80 | 6.36 | DNMT2 | |
| Cn-methylase10 | 1 038 | 9 | 345 | 39 700.18 | 5.27 | DNMT2 | |
| 去甲基化酶 | Cn-demethylase1 | 4 254 | 18 | 1 417 | 160 835.45 | 7.46 | DML3 |
| Demethylase | Cn-demethylase2 | 2 805 | 14 | 934 | 106 836.11 | 8.79 | DML3 |
| Cn-demethylase3 | 2 613 | 15 | 870 | 99 036.82 | 9.20 | DML3 | |
| Cn-demethylase4 | 5 244 | 20 | 1 747 | 194 573.45 | 6.65 | DME | |
| Cn-demethylase5 | 4 707 | 18 | 1 568 | 175 882.89 | 5.96 | ROS1 | |
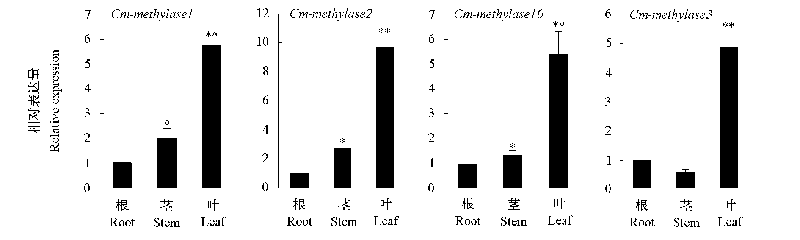
图3 ‘金背大红’菊幼苗期部分DNA甲基转移酶基因和去甲基化酶基因在根、茎和叶的表达分析 将根的表达水平设为1。*表示与其差异显著(P < 0.05),**表示与其差异极显著(P < 0.01)。下同。
Fig. 3 Relative expression of DNA methylases and demethylases gene in root,stem and leaf of Chrysanthemum‘Jinbeidahong’seedling stage Expression leves in root were arbitrarily set to 1. Use * to indicate that the difference is significant(P < 0.05),** to indicate that the difference is significant extremely(P < 0.01). The same below
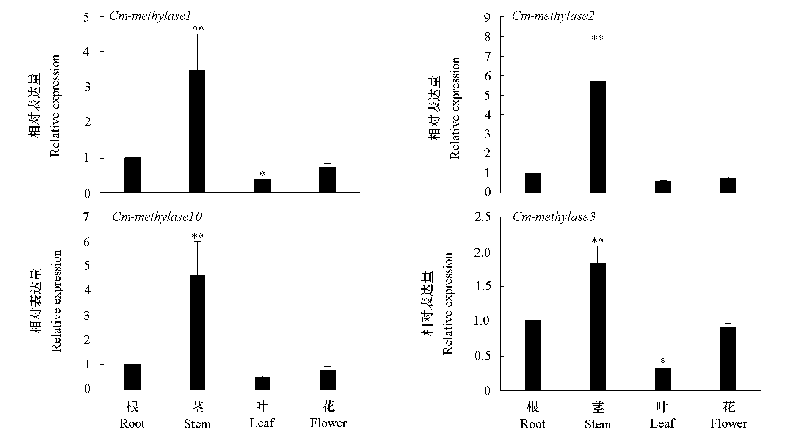
图4 ‘金背大红’菊开花期DNA甲基转移酶基因和去甲基化酶基因在根、茎、叶和花的表达分析
Fig. 4 Relative expression of DNA methylases and demethylases gene in root,stem,leaf and flower of Chrysanthemum‘Jinbeidahong’during flowering stage
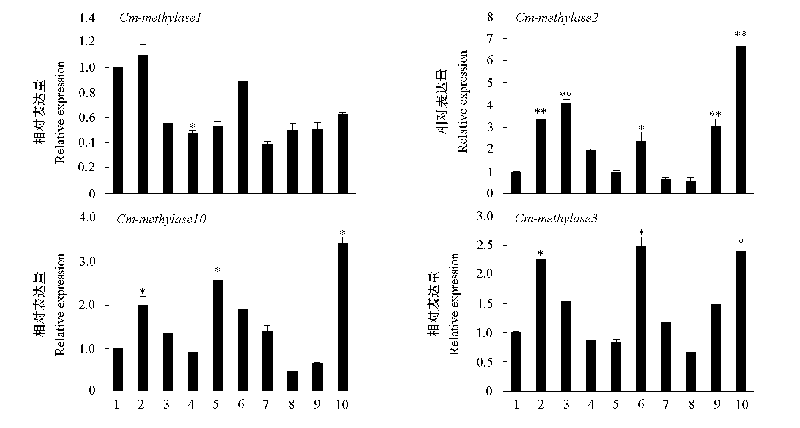
图5 不同品种菊花叶片组织中DNA甲基转移酶基因和去甲基化酶基因的表达分析 1:‘金背大红’;2:‘千丝万缕’;3:‘银龙分水’;4:‘国庆红’;5:‘南农红袖’;6:‘紫精灵’;7:‘杭白菊’;8:‘紫蝶翻飞’;9:‘天女的神秘’;10:‘玉台一号’。将‘金背大红’的表达水平设为1,*表示与其差异显著(P < 0.05),**表示与其差异极显著(P < 0.01)。
Fig. 5 Relative expression of DNA methylases and demethylases in different chrysanthemum cultivars 1:‘Jinbeidahong’;2:‘Qiansiwanlü’;3:‘Yinlongfenshui’;4:‘Guoqinghong’;5:‘Nannong Hongxiu’;6:‘Zijingling’;7:‘Hangbaiju’;8:‘Zidie Fanfei’;9:‘Tiannvdeshenmi’;10:‘Yutaiyihao’. Expression leves of‘Jinbeidahong’were arbitrarily set to 1, use * to indicate that the difference is significant(P < 0.05),** to indicate that the difference is significant extremely(P < 0.01).
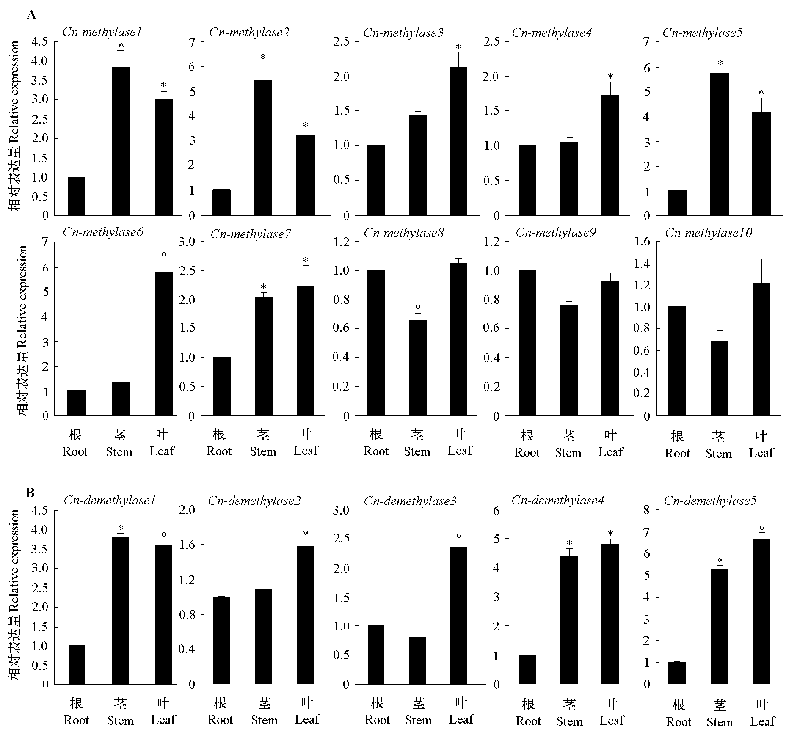
图6 菊花脑幼苗期DNA甲基转移酶(A)和去甲基化酶(B)在根、茎和叶的表达分析
Fig. 6 Expression analysis of DNA methyltransferase(A)and demethylase(B)in root,stem and leaf of Chrysanthemum nankingense seedling stage
| [1] |
Aparicio G, Gotz S, Conesa A, Segrelles D, Blanquer I, Garcia J M, Hernandez V, Robles M, Talon M. 2006. Blast2GO goes grid:developing a grid-enabled prototype for functional genomics analysis. Studies in Health Technology and Informatics, 120:194-204.
pmid: 16823138 |
| [2] | Ashapkin V V, Kutueva L I, Vanyushin B F. 2016. Plant DNA methyltransferase genes:multiplictiy,expression,methylation patterns. Biochemistry, 81 (2):141-151. |
| [3] |
Chan W L, Henderson I R, Jacobsen S E. 2005. Gardening the genome:DNA methylation in Arabidopsis thaliana. Nature Reviews Genetics, 6 (5):351-360.
doi: 10.1038/nrg1601 URL |
| [4] | Chang Lin-lin. 2009. Isolation and expression analysis of DNA methyltransferase genes in strawberry and apple[Ph. D. Dissertation]. Shenyang: Shenyang Agricultural University. (in Chinese) |
| 常琳琳. 2009. 草莓和苹果DNA甲基转移酶基因分离与表达分析[博士论文]. 沈阳: 沈阳农业大学. | |
| [5] | Dai Si-lan, Wang Wen-kui, Huang Jia-ping. 2002. Advances of researches on phylogeny of Dendranthema and origin of chrysanthemum. Journal of Beijing Forestry University, 24 (6):230-234. (in Chinese) |
| 戴思兰, 王文奎, 黄家平. 2002. 菊属系统学及菊花起源的研究进展. 北京林业大学学报, 24 (6):230-234. | |
| [6] |
Dong W, Li M M, Li Z A, Li S L, Zhu Y, Hong X, Wang Z C. 2020. Transcriptome analysis of the molecular mechanism of Chrysanthemum flower color change under short-day photoperiods. Plant Physiology and Biochemistry, 146:315-328.
doi: S0981-9428(19)30492-9 pmid: 31785518 |
| [7] |
Fan S H, Liu H F, Liu J, Hua W, Xu S M, Li J. 2020. Systematic analysis of the DNA methylase and demethylase gene families in rapeseed(Brassica napus L.)and their expression variations after salt and heat stresses. International Journal of Molecular Sciences, 21 (3):953.
doi: 10.3390/ijms21030953 URL |
| [8] | Fu Q S, Song A X, Hu H Y. 2012. Structural aspects of ubiquitin binding specificities. Current Protein & Peptide Science, 13 (5):482-489. |
| [9] |
Furner I J, Matzke M. 2011. Methylation and demethylation of the Arabidopsis genome. Current Opinion in Plant Biology, 14 (2):137-141.
doi: 10.1016/j.pbi.2010.11.004 pmid: 21159546 |
| [10] |
Gong Z Z, Morales-Ruiz T, Ariza R R, Roldan-Arjona T, David L, Zhu J K. 2002. ROS1,a repressor of transcriptional gene silencing in Arabidopsis,encodes a DNA glycosylaselyase. Cell, 111 (6):803-814.
doi: 10.1016/S0092-8674(02)01133-9 URL |
| [11] |
Gu T T, Ren S, Wang Y H, Han Y H, Li Y. 2016. Characterization of DNA methyltransferase and demethylase genes in Fragaria vesca. Molecular Genetics and Genomics, 291 (3):1333-1345.
doi: 10.1007/s00438-016-1187-y URL |
| [12] |
Kang D R, Dai S L, Gao K, Zhang F, Luo H. 2019a. Morphological variation of Chrysanthemum lavandulifolium induced by 5-azaC treatment. Scientia Horticulturae, 257:108645.
doi: 10.1016/j.scienta.2019.108645 URL |
| [13] |
Kang D R, Dai S L, Gao K, Zhang F, Luo H. 2019b. Morphological variation of five cut chrysanthemum cultivars induced by 5-azaC treatment. HortScience, 54 (7):1208-1216.
doi: 10.21273/HORTSCI14012-18 URL |
| [14] | Li J, Li C L, Lu S F. 2018. Identification and characterization of the cytosine-5 DNA methyltransferase gene family in Salvia miltiorrhiza. Peer J, 6:4461. |
| [15] |
Li S L, Li M M, Li Z A, Zhu Y, Ding H X, Fan X X, Li F, Wang Z C. 2019. Effects of the silencing of CmMET1 by RNA interference in chrysanthemum(Chrysanthemum morifolium). Plant Biotechnology Reports, 13 (1):63-72.
doi: 10.1007/s11816-019-00516-5 URL |
| [16] | Liu Yan-hua, Li Zhong-ai, Li Jie, Liu Xiao, Wang Zi-cheng. 2017. Effect of curcumin on DNA methylation and growth of chrysanthemum. Henan Agricultural Sciences, 46 (5):100-105. (in Chinese) |
| 刘艳华, 李忠爱, 李杰, 刘晓, 王子成. 2017. 姜黄素对菊花DNA甲基化及其生长发育的影响. 河南农业科学, 46 (5):100-105. | |
| [17] |
Matzke M A, Mette M F, Matzke A J M. 2000. Transgene silencing by the host genome defense:implications for the evolution of epigenetic control mechanisms in plants and vertebrates. Plant Molecular Biology, 43 (2-3):401-415.
pmid: 10999419 |
| [18] | Mok Y G, Uzawa R, Lee J, Werner G M, Eichman B F, Fischer R L, Huh J H. 2010. Domain structure of the DEMETER 5-methylcytosine DNA glycosylase. Proceedings of the National Academy of Sciences of the United States of America, 107 (45):19225-19230. |
| [19] | Morales-Ruiz T, Ortega-Galisteo A P, Ponferrada-Marin M I, Martinez-Macias M I, Ariza R R, Roldan-Arjona T. 2006. DEMETER and pepressor of silencing 1 encode 5-methylcytosine DNA glycosylases. Proceedings of the National Academy of Sciences of the United States of America, 103 (18):6853-6858. |
| [20] |
Nielsen P R, Nietlispach D, Mott H R, Callaghan J, Bannister A, Kouzarides T, Murzin A G, Murzina N V, Laue E D. 2002. Structure of the HP 1 chromodomain bound to histone H3 methylated at lysine 9. Nature, 416 (6876):103-107.
doi: 10.1038/nature722 URL |
| [21] |
Oliver A W, Jones S A, Roe S M, Matthews S, Goodwin G H, Pearl L H. 2005. Crystal structure of the proximal BAH domain of the polybromo protein. Biochemical Journal, 389 (3):657-664.
doi: 10.1042/BJ20050310 URL |
| [22] |
Ortega-Galisteo A P, Morales-Ruiz T, Ariza R R, Roldan-Arjona, 2008. Arabidopsis DEMETER-LIKE proteins DML2 and DML 3 are required for appropriate distribution of DNA methylation marks. Plant Molecular Biology, 67 (6):671-681.
doi: 10.1007/s11103-008-9346-0 pmid: 18493721 |
| [23] |
Qi X Y, Wang H B, Song A P, Jiang J F, Chen S M, Chen F D. 2018. Genomic and transcriptomic alterations following intergeneric hybridization and polyploidization in the Chrysanthemum nankingense × Tanacetum vulgare hybrid and allopolyploid (Asteraceae). Horticulture Research, 5:5.
doi: 10.1038/s41438-017-0003-0 URL |
| [24] |
Qian Y X, Xi Y L, Cheng B J, Zhu S W. 2014. Genome-wide identification and expression profiling of DNA methyltransferase gene family in maize. Plant Cell Reports, 33 (10):1661-1672.
doi: 10.1007/s00299-014-1645-0 URL |
| [25] |
Silva H G, Sobral R S, Magalhes A P, Morais-Cecilio L, Costa M M R. 2020. Genome-wide identification of epigenetic regulators in Quercus suber L. International Journal of Molecular Sciences, 21 (11):3783.
doi: 10.3390/ijms21113783 URL |
| [26] |
Song C, Liu Y F, Song A P, Dong G Q, Zhao H B, Sun W, Ramakrishnan S, Wang Y, Wang S B, Li T Z, Niu Y, Jiang J F, Dong B, Xia Y, Chen S M, Hu Z G, Chen F D, Chen S L. 2018. The Chrysanthemum nankingense genome provides insights into the evolution and diversification of chrysanthemum flowers and medicinal traits. Molecular Plant, 11 (12):1482-1491.
doi: S1674-2052(18)30308-3 pmid: 30342096 |
| [27] | Wang H B, Qi X Y, Chen S M, Fang W M, Guan Z Y, Teng N J, Liao Y, Jiang J F, Chen F D. 2015. Limited DNA methylation variation and the transcription of MET1 and DDM1 in the genus Chrysanthemum(Asteraceae):following the track of polyploidy. Frontiers in Plant Science, 6:68. |
| [28] | Xu Sheng, Zhang Xin, Liu Decai, Gu Tingting. 2020. Identification and functional analysis of N6-Adenine methylation(6mA)methyltransferase genes in Fragaria vesca. Acta Horticulturae Sinica, 47 (11):2194-2206. (in Chinese) |
| 徐昇, 张鑫, 刘德才, 顾婷婷. 2020. 森林草莓6mA甲基转移酶基因鉴定及功能分析. 园艺学报, 47 (11):2194-2206. | |
| [29] |
Yamauchi T, Johzuka-Hisatomi Y, Terada R, Nakamura I, Lida S. 2014. The MET1b gene encoding a maintenance DNA methyltransferase is indispensable for normal development in rice. Plant Molecular Biology, 85 (3):219-232.
doi: 10.1007/s11103-014-0178-9 pmid: 24535433 |
| [30] |
Yu Yuan-yuan, Guo Yun-hui, Wen Li-zhu, Sun Cui-hui, Fan Hong-mei, Sun Xian-zhi, Wang Wen-li, Sun Xia, Zheng Cheng-shu. 2018. Effects of DNA methylation level on the nitrate uptake of roots in Chrysanthemum morifolium. Plant Physiology Journal, 54 (5):886-894. (in Chinese)
doi: 10.1104/pp.54.6.886 URL |
| 于媛媛, 郭芸珲, 温立柱, 孙翠慧, 樊红梅, 孙宪芝, 王文莉, 孙霞, 郑成淑. 2018. 菊花基因组DNA甲基化水平对根系硝态氮吸收的影响. 植物生理学报, 54 (5):886-894. | |
| [31] |
Zhang C M, Hao Y J. 2020. Advances in genomic,transcriptomic,and metabolomic analyses of fruit quality in fruit crops. Horticultural Plant Journal, 6 (6):361-371.
doi: 10.1016/j.hpj.2020.11.001 URL |
| [32] |
Zhang F, Deng C Y, Dai S L. 2020. MSAP analysis reveals the DNA methylation dynamics during capitulum development in Chrysanthemum lavandulifolium. Canadian Journal of Plant Science, 101 (3):1-31.
doi: 10.1139/cjps-2019-0318 URL |
| [33] |
Zhang H M, Lang Z B, Zhu J K. 2018. Dynamics and function of DNA methylation in plants. Nature Reviews Molecular Cell Biology, 19 (8):489-506.
doi: 10.1038/s41580-018-0016-z URL |
| [34] | Zhou Chenping, Yang Min, Guo Jinju, Kuang Ruibin, Yang Hu, Huang Bingxiong, Wei Yuerong. 2022. Dynamic changes in DNA methylome and transcriptome patterns during papaya fruit ripening. Acta Horticulturae Sinica, 49 (3):519-532. (in Chinese) |
| 周陈平, 杨敏, 郭金菊, 邝瑞彬, 杨护, 黄炳雄, 魏岳荣. 2022. 番木瓜成熟过程中全基因组DNA甲基化和转录组变化分析. 园艺学报, 49 (3):519-532. |
| [1] | 王晓晨, 聂子页, 刘先菊, 段 伟, 范培格, 梁振昌, . 脱落酸对‘京香玉’葡萄果实单萜物质合成的影响[J]. 园艺学报, 2023, 50(2): 237-249. |
| [2] | 任 菲, 卢苗苗, 刘吉祥, 陈信立, 刘道凤, 眭顺照, 马 婧. 蜡梅胚胎晚期丰富蛋白基因CpLEA的表达及抗性分析[J]. 园艺学报, 2023, 50(2): 359-370. |
| [3] | 翟含含, 翟宇杰, 田义, 张叶, 杨丽, 温陟良, 陈海江. 桃SAUR家族基因分析及PpSAUR5功能鉴定[J]. 园艺学报, 2023, 50(1): 1-14. |
| [4] | 赵雪艳, 王琪, 王莉, 王方圆, 王庆, 李艳. 基于比较转录组的延胡索组织差异性表达分析[J]. 园艺学报, 2023, 50(1): 177-187. |
| [5] | 邵凤清, 罗秀荣, 王奇, 张宪智, 王文彩. 果实成熟过程中的DNA甲基化调控研究进展[J]. 园艺学报, 2023, 50(1): 197-208. |
| [6] | 叶志琴, 杨 娟. 菊花新品种‘紫鬓云’[J]. 园艺学报, 2022, 49(S2): 173-174. |
| [7] | 赵艳莉, 李战鸿, 曹 琴, 戴妙飞, 高萌萌, 孙珍珠, 李会宽, 李永华, 卞书迅, 黄 淦. 菊花新品种‘汴京庆典黄’[J]. 园艺学报, 2022, 49(S2): 175-176. |
| [8] | 曹 琴, 赵艳莉, 李战鸿, 高萌萌, 戴妙飞, 李永华, 李会宽, 孙珍珠, 卞书迅, 李 菲. 小菊新品种‘汴京紫精灵’[J]. 园艺学报, 2022, 49(S2): 177-178. |
| [9] | 高彦龙, 吴玉霞, 张仲兴, 王双成, 张瑞, 张德, 王延秀. 苹果ELO家族基因鉴定及其在低温胁迫下的表达分析[J]. 园艺学报, 2022, 49(8): 1621-1636. |
| [10] | 邱子文, 刘林敏, 林永盛, 林晓洁, 李永裕, 吴少华, 杨超. 千层金MbEGS基因的克隆与功能分析[J]. 园艺学报, 2022, 49(8): 1747-1760. |
| [11] | 陶鑫, 朱荣香, 贡鑫, 吴磊, 张绍铃, 赵建荣, 张虎平. 梨果糖激酶基因PpyFRK5在果实蔗糖积累中的作用[J]. 园艺学报, 2022, 49(7): 1429-1440. |
| [12] | 郑林, 王帅, 刘语诺, 杜美霞, 彭爱红, 何永睿, 陈善春, 邹修平. 柑橘响应黄龙病菌侵染的NAC基因的克隆及表达分析[J]. 园艺学报, 2022, 49(7): 1441-1457. |
| [13] | 张秋悦, 刘昌来, 于晓晶, 杨甲定, 封超年. 盐胁迫条件下杜梨叶片差异表达基因qRT-PCR内参基因筛选[J]. 园艺学报, 2022, 49(7): 1557-1570. |
| [14] | 马维峰, 李艳梅, 马宗桓, 陈佰鸿, 毛娟. 苹果POD家族基因的鉴定与MdPOD15的功能分析[J]. 园艺学报, 2022, 49(6): 1181-1199. |
| [15] | 张凯, 麻明英, 王萍, 李益, 金燕, 盛玲, 邓子牛, 马先锋. 柑橘HSP20家族基因鉴定及其响应溃疡病菌侵染表达分析[J]. 园艺学报, 2022, 49(6): 1213-1232. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司