
园艺学报 ›› 2022, Vol. 49 ›› Issue (3): 519-532.doi: 10.16420/j.issn.0513-353x.2020-1046
周陈平1, 杨敏1, 郭金菊2, 邝瑞彬1, 杨护1, 黄炳雄1, 魏岳荣1,*( )
)
收稿日期:2021-07-19
修回日期:2021-08-20
出版日期:2022-03-25
发布日期:2022-03-25
通讯作者:
魏岳荣
E-mail:weid18@163.com
基金资助:
ZHOU Chenping1, YANG Min1, GUO Jinju2, KUANG Ruibin1, YANG Hu1, HUANG Bingxiong1, WEI Yuerong1,*( )
)
Received:2021-07-19
Revised:2021-08-20
Online:2022-03-25
Published:2022-03-25
Contact:
WEI Yuerong
E-mail:weid18@163.com
摘要:
采用全基因组DNA甲基化(WGBS)和转录组测序(RNA-seq)技术对番木瓜4个成熟时期(绿熟期、破色期、熟化期、腐熟期)的果实样品进行高通量测序。C位点、CG、CHG和CHH序列平均甲基化水平分别为17.15%、49.33%、34.30%和8.28%。全基因组DNA甲基化水平在果实成熟过程中持续下降,同时差异表达基因(DEG)数量逐渐上升。全基因组DNA甲基化和转录组数据联合分析表明,基因表达水平与启动子及基因体甲基化水平呈显著正相关。筛选出CpACO4、CpPDS、CpXTH30和CpXTH31等4个番木瓜果实成熟差异表达候选基因,利用荧光定量PCR和McrBC-PCR分别检测其表达水平和相应DMR序列甲基化水平,结果分别同转录组和全基因组DNA甲基化一致,推测这些基因在番木瓜果实成熟过程中起重要作用。
中图分类号:
周陈平, 杨敏, 郭金菊, 邝瑞彬, 杨护, 黄炳雄, 魏岳荣. 番木瓜成熟过程中全基因组DNA甲基化和转录组变化分析[J]. 园艺学报, 2022, 49(3): 519-532.
ZHOU Chenping, YANG Min, GUO Jinju, KUANG Ruibin, YANG Hu, HUANG Bingxiong, WEI Yuerong. Dynamic Changes in DNA Methylome and Transcriptome Patterns During Papaya Fruit Ripening[J]. Acta Horticulturae Sinica, 2022, 49(3): 519-532.
| 用途 Application | 基因名称 Gene name | 上游引物(5′-3′) Forward primer | 下游引物(5′-3′) Reverse primer |
|---|---|---|---|
| McrBC-PCR | CpXTH30 | GGTTGGCTAATGATTGT | AGAAGGAAGGAGGGTGC |
| CpPDS | CTAGTTTCTAACTGCGTAT | AACCATTATCATGTGGG | |
| CpACO4 | GAAGACCATTAGCCACA | TTCGCTTAACTTAAACAC | |
| CpXTH31 | ATGAAGTAGGGTGAGAA | ATGGTGGAATAAGTGTT | |
| qRT-PCR | CpXTH30 | GTTCCATCGGTATAGCAT | CTTTGTATCTTCCACCTGA |
| CpPDS | AGTTGGCGTCCCTGTTA | GGATTGATTTGGGTTGTA | |
| CpACO4 | CACGAGCAGCCAAACGA | TCACCCACCCATCCCAC | |
| CpXTH31 | GGGAACCCTACACTTTG | CTTGGATACCTCCGAAT |
表1 McrBC-PCR和实时定量PCR引物
Table 1 Primers used for McrBC-PCR and qRT-PCR
| 用途 Application | 基因名称 Gene name | 上游引物(5′-3′) Forward primer | 下游引物(5′-3′) Reverse primer |
|---|---|---|---|
| McrBC-PCR | CpXTH30 | GGTTGGCTAATGATTGT | AGAAGGAAGGAGGGTGC |
| CpPDS | CTAGTTTCTAACTGCGTAT | AACCATTATCATGTGGG | |
| CpACO4 | GAAGACCATTAGCCACA | TTCGCTTAACTTAAACAC | |
| CpXTH31 | ATGAAGTAGGGTGAGAA | ATGGTGGAATAAGTGTT | |
| qRT-PCR | CpXTH30 | GTTCCATCGGTATAGCAT | CTTTGTATCTTCCACCTGA |
| CpPDS | AGTTGGCGTCCCTGTTA | GGATTGATTTGGGTTGTA | |
| CpACO4 | CACGAGCAGCCAAACGA | TCACCCACCCATCCCAC | |
| CpXTH31 | GGGAACCCTACACTTTG | CTTGGATACCTCCGAAT |
| 成熟期 Stage | 序列总数量 Total reads | 对比参考基 因组获得的 序列数量 Mapped reads | 匹配率/% Mapping rate | 重复序列 占比/% Duplication rate | ≥ Q30/% | 测序深度 Sequencing depth | 甲基化C位点 数量Number of Methylated C site | 平均甲基化水平/% Mean methylation level | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| C | CG | CHG | CHH | ||||||||
| 绿熟期 Green | 49 375 154 | 35 135 889 | 70.97 | 14.37 | 92.78 | 21.87 | 251 784 698 | 17.79 | 51.22 | 35.59 | 8.62 |
| 破色期 Color break | 49 361 724 | 34 881 444 | 70.67 | 12.75 | 91.56 | 22.08 | 258 718 931 | 17.51 | 50.64 | 35.09 | 8.44 |
| 熟化期 Half yellow | 49 461 069 | 34 774 513 | 70.36 | 12.62 | 92.57 | 21.95 | 232 054 629 | 17.04 | 48.60 | 33.76 | 8.26 |
| 腐熟期 Full yellow | 48 863 321 | 34 071 375 | 70.61 | 10.79 | 92.19 | 22.54 | 233 578 504 | 16.26 | 46.88 | 32.77 | 7.81 |
表2 番木瓜果实不同成熟时期全基因组DNA甲基化测序数据统计情况
Table 2 Summary of WGBS data of papaya fruits at different ripening stages
| 成熟期 Stage | 序列总数量 Total reads | 对比参考基 因组获得的 序列数量 Mapped reads | 匹配率/% Mapping rate | 重复序列 占比/% Duplication rate | ≥ Q30/% | 测序深度 Sequencing depth | 甲基化C位点 数量Number of Methylated C site | 平均甲基化水平/% Mean methylation level | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| C | CG | CHG | CHH | ||||||||
| 绿熟期 Green | 49 375 154 | 35 135 889 | 70.97 | 14.37 | 92.78 | 21.87 | 251 784 698 | 17.79 | 51.22 | 35.59 | 8.62 |
| 破色期 Color break | 49 361 724 | 34 881 444 | 70.67 | 12.75 | 91.56 | 22.08 | 258 718 931 | 17.51 | 50.64 | 35.09 | 8.44 |
| 熟化期 Half yellow | 49 461 069 | 34 774 513 | 70.36 | 12.62 | 92.57 | 21.95 | 232 054 629 | 17.04 | 48.60 | 33.76 | 8.26 |
| 腐熟期 Full yellow | 48 863 321 | 34 071 375 | 70.61 | 10.79 | 92.19 | 22.54 | 233 578 504 | 16.26 | 46.88 | 32.77 | 7.81 |
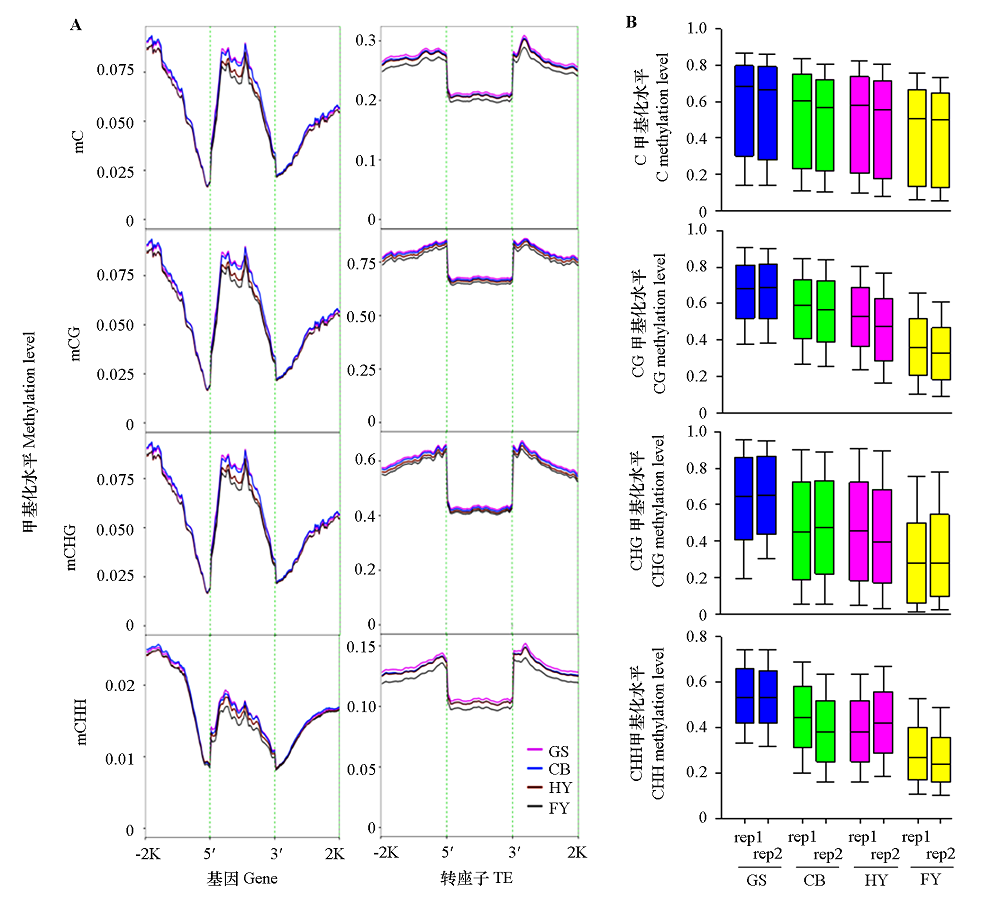
图2 不同成熟期番木瓜果实的基因(gene)和转座子(TE)DNA甲基化水平(A)和C、CG、CHG和CHH序列的甲基化水平(B) GS:绿熟期;CB:破色期;HY:熟化期;FY:腐熟期;-2K和2K:基因上游和下游2 kb区域;5′和3′:基因5′和3′侧翼区域;rep1和rep2:2个生物学重复。下同。
Fig. 2 DNA methylation levels of genes and TEs(A)and C,CG,CHG,and CHH contexts(B)at different ripening stages of papaya fruits GS:Green stage;CB:Color break stage;HY:Half yellow stage;FY:Full yellow stage;-2K and 2K:2 kb upstream and downstream regions of genes;5′ and 3′:5′ and 3′ flanking regions of genes;rep1 and rep2:Two biological replicates. The same below.
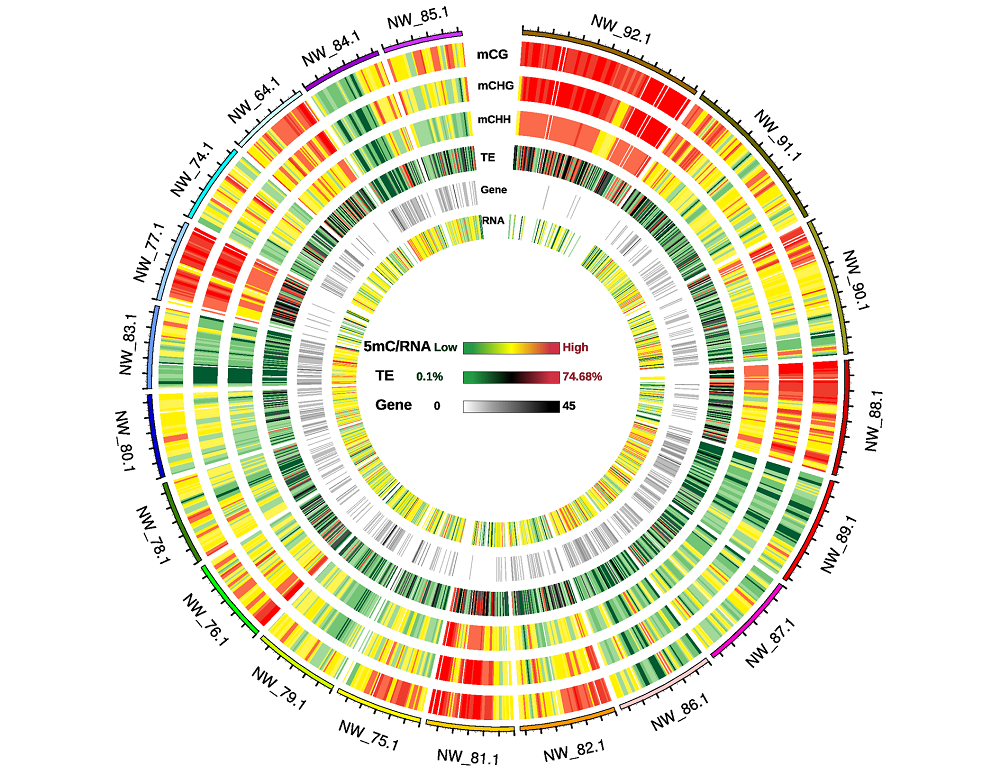
图4 20条基因组骨架甲基化密度circos图 图谱外圈编号:20条基因组骨架的编号;5mC/RNA、TE和Gene热标:分别为CG、CHG、CHH序列甲基化密度、RNA丰度、转座子重复序列比例和基因数量由低到高。
Fig. 4 Circos plot of twenty genome scaffold methylation density Number on outer rim:Number of the twenty genome scaffolds;Scales of 5mC/RNA,TE and Gene:The methylation levels of CG,CHG,CHH sequence context,RNA abundance,the ratio of transposon repeats and gene number were from low to high,respectively.
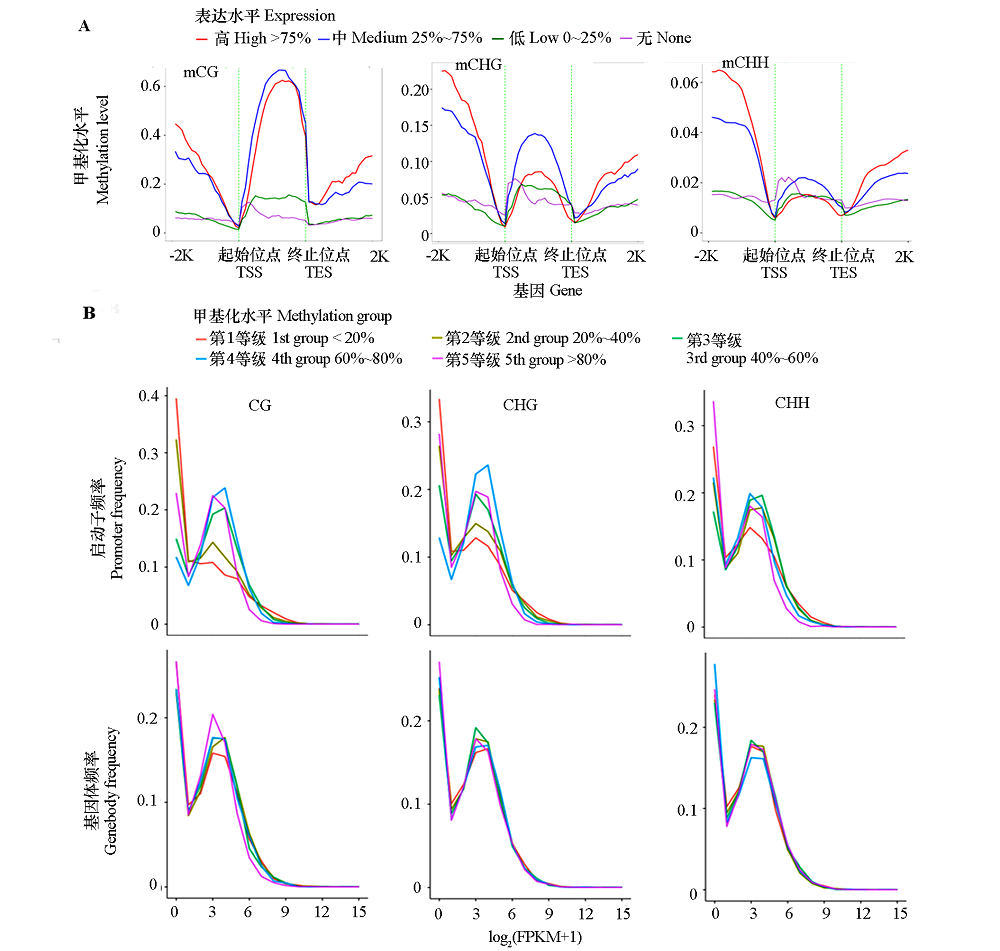
图5 3种序列甲基化水平在基因体及其上、下游2 kb区域的分布(A)和基因体及启动子甲基化水平与基因表达水平关系(B) -2K和2K:基因上游和下游2 kb区域;表达水平:FPKM值 < 1基因的FPKM值为无(不表达),FPKM值 ≥ 1基因的FPKM值由小到大排列,求得上下四分位处的FPKM值;甲基化水平等级:基因体或启动子甲基化水平值由小到大排列,求得第20/40/60/80百分位处的甲基化水平值。
Fig. 5 Relationship between distribution of methylation levels in three contexts within the genebody,upstream,and downstream 2 kb rigions(A)and methylation levels of genebody and promoterwith the gene expression levels(B) -2K and 2K:2 kb upstream and downstream regions of genes;Expression:genes with FPKM value < 1 were considered as none(non-expressed),expressed genes(FPKM value ≥ 1)were ranked from small to large and divided into three groups based on the upper and lower quartile;Methylation group:Methylation levels of genebodies or promoters were ranked from small to large and divided into quinitiles.
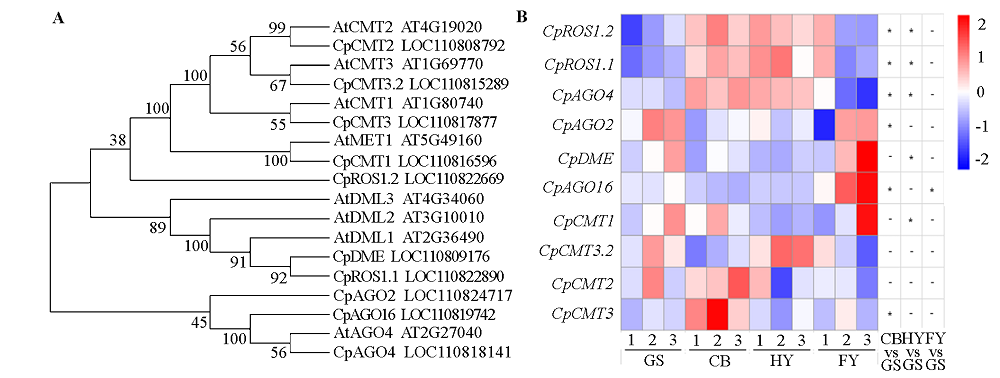
图7 番木瓜DNA甲基化基因的同源比对(A)及其在不同成熟时期表达变化(B) 热标:由蓝到红表示基因表达量由低到高。1 ~ 3:3个生物学重复。*:P < 0.05;-:无显著差异。
Fig. 7 Homologous alignment of DNA methylation genes(A)and their expression changes at different ripening stages in papaya fruits(B) Scale:From blue to red represent the gene expression levels was from low to high. 1-3:Three biological replicates.*:P < 0.05;-:No significant difference.
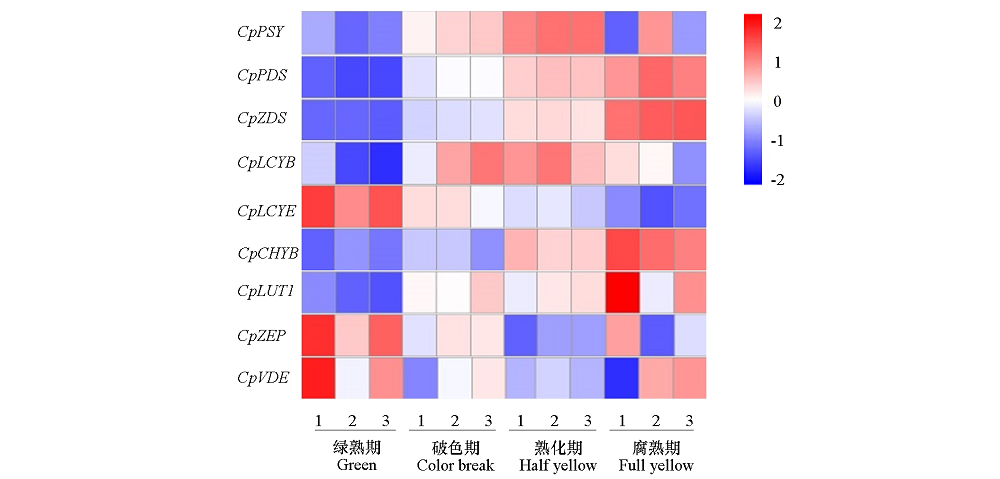
图8 类胡萝卜素生物合成基因在番木瓜果实不同成熟时期的表达变化 热标:由蓝到红表示基因表达量由低到高;1 ~ 3:3个生物学重复。
Fig. 8 Expression changes of carotenoid biosynthetic-related genes at different ripening stages of papaya fruits Scale:From blue to red represent the gene expression levels was from low to high;1-3:Three biological replicates.
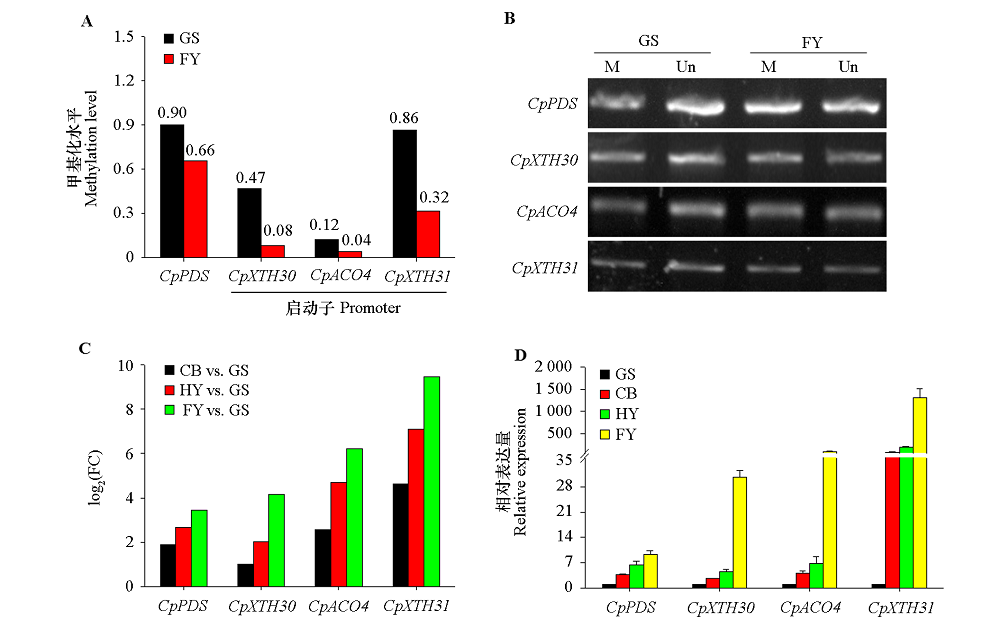
图9 4个候选基因DMR甲基化水平及其在番木瓜不同成熟时期的表达分析 A:CpACO4、CpXTH30和CpXTH31启动子及CpPDS基因体DMR甲基化水平;B:McrBC-PCR验证(M:McrBC酶切的PCR产物,Un:未被酶切的PCR产物);C:表达倍数(FC);D:qRT-PCR验证。
Fig. 9 The DMRs methylation and expression levels of four candidate genes at different maturation stages of papaya fruits A:DMRs methylation levels of CpACO4,CpXTH30 and CpXTH31 promoters and CpPDS genebody;B:The McrBC-PCR validation(M:PCR products been digested by McrBC,Un:PCR products not been digested);C:Fold change(FC);D:qRT-PCR validation.
| [1] |
Ahmad A, Zhang Y, Cao X F. 2010. Decoding the epigenetic language of plant development. Molecular Plant, 3(4):719-728.
doi: 10.1093/mp/ssq026 pmid: 20663898 |
| [2] |
Bolger A M, Lohse M, Usadel B. 2014. Trimmomatic:a flexible trimmer for Illumina sequence data. Bioinformatics, 30(15):2114-2120.
doi: 10.1093/bioinformatics/btu170 URL |
| [3] |
Bonasio R, Tu S, Reinberg D. 2010. Molecular signals of epigenetic states. Science, 330(6004):612-616.
doi: 10.1126/science.1191078 URL |
| [4] | Chen Duanfen, Peng Zhenhua, Gao Zhimin. 2008. Cloning and expression analysis of PDS gene in Narcissus tazetta var. chinensis. Molecular Plant Breeding, 6(3):574-578. (in Chinese) |
| 陈段芬, 彭镇华, 高志民. 2008. 中国水仙八氢番茄红素脱氢酶基因(PDS)的克隆及表达分析. 分子植物育种, 6(3):574-578. | |
| [5] | Chen Yong-ping, Gao Feng, Shen Yan-hong, Zhao Wan-wan, Chen Gui-xin, Wang Ping. 2019. The analysis of ERFs related to fruit ripening in papaya. Acta Horticulturae Sinica, 46(2):252-264. (in Chinese) |
| 陈永萍, 高峰, 申艳红, 赵弯弯, 陈桂信, 王平. 2019. 番木瓜ERF家族与果实成熟相关成员的分析. 园艺学报, 46(2):252-264. | |
| [6] |
Cheng J, Niu Q, Zhang B, Chen K, Yang R, Zhu J K, Zhang Y, Lang Z. 2018. Downregulation of RdDM during strawberry fruit ripening. Genome Biology, 19(1):212.
doi: 10.1186/s13059-018-1587-x URL |
| [7] |
Ding X, Zhu X, Ye L, Xiao S, Wu Z, Chen W, Li X. 2019. The interaction of CpEBF1 with CpMADSs is involved in cell wall degradation during papaya fruit ripening. Horticulture Research, 6(1):13.
doi: 10.1038/s41438-018-0095-1 URL |
| [8] | Du Xiaoyun, Song Laiqing, Zhao Lingling, Liu Meiying, Tang Yan, Sun Yanxia, Jiang Zhongwu, Shu Huairui. 2019. DNA methylation in Red Fuji apple bud sports lines. Acta Horticulturae Sinica, 46(1):107-120. (in Chinese) |
| 杜晓云, 宋来庆, 赵玲玲, 刘美英, 唐岩, 孙燕霞, 姜中武, 束怀瑞. 2019. 红富士苹果芽变系DNA甲基化研究. 园艺学报, 46(1):107-120. | |
| [9] |
Fabi J P, Do Prado S B R. 2019. Fast and furious:ethylene-triggered changes in the metabolism of papaya fruit during ripening. Frontiers in Plant Science, 10:535.
doi: 10.3389/fpls.2019.00535 URL |
| [10] | Fan Zhong-qi, Kuang Jian-fei, Lu Wang-jin, Chen Jian-ye. 2015. Advances in research of the mechanism of transcription factors involving in regulating fruit ripening and senescence. Acta Horticulturae Sinica, 42(9):1649-1663. (in Chinese) |
| 范中奇, 邝健飞, 陆旺金, 陈建业. 2015. 转录因子调控果实成熟和衰老机制研究进展. 园艺学报, 42(9):1649-1663. | |
| [11] |
Finnegan E J, Kovac K A. 2000. Plant DNA methyltransferases. Plant Molecular Biology, 43:189-201.
pmid: 10999404 |
| [12] | Grafi G. 2011. Epigenetics in plant development and response to stress. Biochimica et Biophysica Acta, 1809:351-352. |
| [13] |
He X J, Chen T, Zhu J K. 2011. Regulation and function of DNA methylation in plants and animals. Cell Research, 21:442-465.
doi: 10.1038/cr.2011.23 URL |
| [14] | Huang H, Liu R, Niu Q, Tang K, Zhang B, Zhang H, Chen K, Zhu J K, Lang Z. 2019. Global increase in DNA methylation during orange fruit development and ripening. Proceedings of the National Academy of Sciences of the United States of America, 116(4):1430-1436. |
| [15] | Huang Wen-jun, Zhong Cai-hong. 2017. Research advances in the postharvest physiology of kiwifruit. Plant Science Journal, 35(4):622-630. (in Chinese) |
| 黄文俊, 钟彩虹. 2017. 猕猴桃果实采后生理研究进展. 植物科学学报, 35(4):622-630. | |
| [16] |
Krueger F, Andrews S R. 2011. Bismark:a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics, 27(11):1571-1572.
doi: 10.1093/bioinformatics/btr167 pmid: 21493656 |
| [17] |
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones S J, Marra M A. 2009. Circos:an information aesthetic for comparative genomics. Genome Research, 19:1639-1645.
doi: 10.1101/gr.092759.109 pmid: 19541911 |
| [18] | Lang Z, Wang Y, Tang K, Tang D, Datsenka T, Cheng J, Zhang Y, Handa A K, Zhu J K. 2017. Critical roles of DNA demethylation in the activation of ripeninginduced genes and inhibition of ripening-repressed genes in tomato fruit. Proceedings of the National Academy of Sciences of the United States of America, 114:E4511-E4519. |
| [19] |
Law J A, Jacobsen S E. 2010. Establishing,maintaining and modifying DNA methylation patterns in plants and animals. Nature Reviews Genetics, 11:204-220.
doi: 10.1038/nrg2719 URL |
| [20] |
Li X, Zhu J, Hu F, Ge S, Ye M, Xiang H, Zhang G, Zheng X, Zhang H, Zhang S, Li Q, Luo R, Yu C, Yu J, Sun J, Zou X, Cao X, Xie X, Wang J, Wang W. 2012. Single-base resolution maps of cultivated and wild rice methylomes and regulatory roles of DNA methylation in plant gene expression. BMC Genomics, 13:300.
doi: 10.1186/1471-2164-13-300 URL |
| [21] |
Lister R, O’Malley R C, Tonti-Filippini J, Gregory B D, Berry C C, Millar A H, Ecker J R. 2008. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell, 133:523-536.
doi: 10.1016/j.cell.2008.03.029 pmid: 18423832 |
| [22] | Liu R, How-Kit A, Stammitti L, Teyssier E, Rolin D, Mortain-Bertrand A, Halle S, Liu M, Kong J, Wu C, Degraeve-Guibault C, Chapman N H, Maucourt M, Hodgman T C, Tost J, Bouzayen M, Hong Y, Seymour G B, Giovannoni J J, Gallusci P. 2015. A DEMETER-like DNA demethylase governs tomato fruit ripening. Proceedings of the National Academy of Sciences of the United States of America, 112:10804-10809. |
| [23] | Liu Xin, Chen Yunzhu, Kim Pyol, Kim Min-Jun, Song Hyondok, Li Yuhua, Wang Yu. 2020. Progress on molecular mechanism and regulation of tomato fruit color formation. Acta Horticulturae Sinica, 47(9):1689-1704. (in Chinese) |
| 刘昕, 陈韵竹, Kim Pyol, Kim Min-Jun, Song Hyondok, 李玉花, 王宇. 2020. 番茄果实颜色形成的分子机制及调控研究进展. 园艺学报, 47(9):1689-1704. | |
| [24] |
Lu X, Wang X, Chen X, Shu N, Wang J, Wang D, Wang S, Fan W, Guo L, Guo X, Ye W. 2017. Single-base resolution methylomes of upland cotton(Cossypium hirsutum L.)reveal epigenome modification in response to drought stress. BMC Genomics, 18(1):297.
doi: 10.1186/s12864-017-3681-y URL |
| [25] |
Manning K, Tör M, Poole M, Hong Y, Thompson A J, King G J, Giovannoni J J, Seymour G B. 2006. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nature Genetics, 38(8):948.
doi: 10.1038/ng1841 URL |
| [26] |
Marty I, Bureau S, Sarkissian G, Gouble B, Audergon J M, Albagnac G. 2005. Ethylene regulation of carotenoid accumulation and carotenogenic gene expression in colour-contrasted apricot varieties(Prunus armeniaca). Journal of Experimental Botany, 56(417):1877-1886.
pmid: 15911563 |
| [27] |
Muñoz-Bertomeu J, Miedes E, Lorences E P. 2013. Expression of xyloglucan endotransglucosylase/hydrolase(XTH)genes and XET activity in ethylene treated apple and tomato. Journal of Plant Physiology, 170(13):1194-1201.
doi: 10.1016/j.jplph.2013.03.015 pmid: 23628624 |
| [28] |
Narsaiah K, Wilson R A, Gokul K, Mandge H M, Jha S N, Bhadwal S, Anurag R K, Malik RK, Vij S. 2015. Effect of bacteriocin-incorporated alginate coating on shelf-life of minimally processed papaya(Carica papaya L.). Postharvest Biology and Technology, 100:212-218.
doi: 10.1016/j.postharvbio.2014.10.003 URL |
| [29] |
Niederhuth C E, Bewick A J, Ji L, Alabady M S, Kim K D, Li Q, Rohr N A, Rambani A, Burke J M, Udall J A, Egesi C, Schmutz J, Grimwood J, Jackson S A, Springer N M, Schmitz R J. 2016. Widespread natural variation of DNA methylation within angiosperms. Genome Biology, 17:194.
pmid: 27671052 |
| [30] | Pesaresi P, Mizzotti C, Colombo M, Masiero S. 2014. Genetic regulation and structural changes during tomato fruit development and ripening. Frontiers in Plant Science, 23(5):124. |
| [31] |
Pfaffl M W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research, 29(9):e45.
doi: 10.1093/nar/29.9.e45 pmid: 11328886 |
| [32] |
Qi W, Wang H, Zhou Z, Yang P, Wu W, Li Z, Li X. 2020. Ethylene emission as a potential indicator of Fuji apple flavor quality evaluation under low temperature. Horticultural Plant Journal, 6(4):231-239.
doi: 10.1016/j.hpj.2020.03.007 URL |
| [33] | Seymour DK, Koenig D, Hagmann J, Becker C, Weigel D. 2014. Evolution of DNA methylation patterns in the Brassicaceae is driven by differences in genome organization. PLoS Genetics, 10:e1004785. |
| [34] |
Shen Y H, Lu B G, Feng L, Yang F Y, Geng J J, Ming R, Chen X J. 2017. Isolation of ripening-related genes from ethylene/1-MCP treated papaya through RNA-seq. BMC Genomics, 18(1):671.
doi: 10.1186/s12864-017-4072-0 URL |
| [35] |
Tang K, Lang Z, Zhang H, Zhu J K. 2016. The DNA demethylase ROS1 targets genomic regions with distinct chromatin modifications. Nature Plants, 2:16169.
doi: 10.1038/nplants.2016.169 pmid: 27797352 |
| [36] |
Tian Shi-ping. 2013. Molecular mechanisms of fruit ripening and senescence. Chinese Bulletin of Botany, 48(5):481-488. (in Chinese)
doi: 10.3724/SP.J.1259.2013.00481 URL |
| 田世平. 2013. 果实成熟和衰老的分子调控机制. 植物学报, 48(5):481-488. | |
| [37] | Wang Zhong-feng. 2009. Research advancement in relation of enzymes for cell wall metabolism with fruit softening. Chinese Agricultural Science Bulletin, 25(18):126-130. (in Chinese) |
| 王中凤. 2009. 细胞壁分解酶与果实软化的关系研究进展. 中国农学通报, 25(18):126-130. | |
| [38] |
Xu J, Wang X, Cao H, Xu H, Xu Q, Deng X. 2017. Dynamic changes in methylome and transcriptome patterns in response to methyltransferase inhibitor 5-azacytidine treatment in citrus. DNA Research, 24(5):509-522.
doi: 10.1093/dnares/dsx021 URL |
| [39] |
Xu J, Zhou S, Gong X, Song Y, van Nocker S, Ma F, Guan Q. 2018. Single-base methylome analysis reveals dynamic epigenomic differences associated with water deficit in apple. Plant Biotechnology Journal, 16:672-687.
doi: 10.1111/pbi.2018.16.issue-2 URL |
| [40] | Xu Sheng, Zhang Xin, Liu Decai, Gu Tingting. 2020. Identification and functional analysis of N6-adenine methylation(6mA)methyltransferase genes in Fragaria vesca. Acta Horticulturae Sinica, 47(11):2194-2206. (in Chinese) |
| 徐昇, 张鑫, 刘德才, 顾婷婷. 2020. 森林草莓6mA甲基转移酶基因鉴定及功能分析. 园艺学报, 47(11):2194-2206. | |
| [41] |
Zhang M, Zhang X, Guo L, Qi T, Liu G, Feng J, Shahzad K, Zhang B, Li X, Wang H, Tang H, Qiao X, Wu J, Xing C. 2020. Single-base resolution methylome of cotton cytoplasmic male sterility system reveals epigenomic changes in response to high-temperature stress during anther development. Journal of Experimental Botany, 71:951-969.
doi: 10.1093/jxb/erz470 pmid: 31639825 |
| [42] |
Zhang X. 2008. The epigenetic landscape of plants. Science, 320(5875):489-492.
doi: 10.1126/science.1153996 URL |
| [43] |
Zhong S, Fei Z, Chen Y R, Zheng Y, Huang M, Vrebalov J, McQuinn R, Gapper N, Liu B, Xiang J, Shao Y, Giovannoni J J. 2013. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nature Biotechnology, 31(2):154-159.
doi: 10.1038/nbt.2462 URL |
| [44] | Zhou C, Yang M, Luo X, Kuang R, Yang H, Yao J, Huang B, Wei Y. 2021. Transcriptomic analysis of papaya(Carica papaya L.)shoot explants obtained by leaf- and stem-inoculation methods for adventitious roots induction. Scientia Horticulturae, 276:109762. |
| [45] |
Zhu J K. 2009. Active DNA demethylation mediated by DNA glycosylases. Annual Review of Genetics, 43:143-66.
doi: 10.1146/genet.2009.43.issue-1 URL |
| [1] | 王晓晨, 聂子页, 刘先菊, 段 伟, 范培格, 梁振昌, . 脱落酸对‘京香玉’葡萄果实单萜物质合成的影响[J]. 园艺学报, 2023, 50(2): 237-249. |
| [2] | 张 欣, 漆艳香, 曾凡云, 王艳玮, 谢培兰, 谢艺贤, 彭 军. 香蕉枯萎病菌Dicer-like基因的功能分析[J]. 园艺学报, 2023, 50(2): 279-294. |
| [3] | 任 菲, 卢苗苗, 刘吉祥, 陈信立, 刘道凤, 眭顺照, 马 婧. 蜡梅胚胎晚期丰富蛋白基因CpLEA的表达及抗性分析[J]. 园艺学报, 2023, 50(2): 359-370. |
| [4] | 于婷婷, 李 欢, 宁源生, 宋建飞, 彭璐琳, 贾竣淇, 张玮玮, 杨洪强. 苹果GRAS全基因组鉴定及其对生长素的响应分析[J]. 园艺学报, 2023, 50(2): 397-409. |
| [5] | 翟含含, 翟宇杰, 田义, 张叶, 杨丽, 温陟良, 陈海江. 桃SAUR家族基因分析及PpSAUR5功能鉴定[J]. 园艺学报, 2023, 50(1): 1-14. |
| [6] | 杨植, 张川疆, 杨芯芳, 董梦怡, 王振磊, 闫芬芬, 吴翠云, 王玖瑞, 刘孟军, 林敏娟. 枣与酸枣杂交后代果实遗传倾向及混合遗传分析[J]. 园艺学报, 2023, 50(1): 36-52. |
| [7] | 崔建, 钟雄辉, 刘泽慈, 陈登辉, 李海龙, 韩睿, 乐祥庆, 康俊根, 王超. 结球甘蓝染色体片段替换系构建[J]. 园艺学报, 2023, 50(1): 65-78. |
| [8] | 何成勇, 赵晓丽, 许腾飞, 高德航, 李世访, 王红清. 草莓病毒1山东分离物全基因组分析[J]. 园艺学报, 2023, 50(1): 153-160. |
| [9] | 罗海林, 袁雷, 翁华, 闫佳会, 郭青云, 王文清, 马新明. 蚕豆萎蔫病毒2号青海辣椒分离物的鉴定与全基因组序列克隆[J]. 园艺学报, 2023, 50(1): 161-169. |
| [10] | 忽靖宇, 阙开娟, 缪田丽, 吴少政, 王田田, 张磊, 董鲜, 季鹏章, 董家红. 侵染鸢尾的番茄斑萎病毒鉴定[J]. 园艺学报, 2023, 50(1): 170-176. |
| [11] | 邵凤清, 罗秀荣, 王奇, 张宪智, 王文彩. 果实成熟过程中的DNA甲基化调控研究进展[J]. 园艺学报, 2023, 50(1): 197-208. |
| [12] | 蒋亚君, 陈佳佳, 谭彬, 郑先波, 王伟, 张郎郎, 程钧, 冯建灿. 桃PpIDD11调控花发育的功能研究[J]. 园艺学报, 2022, 49(9): 1841-1852. |
| [13] | 王燕, 刘针杉, 张静, 杨鹏飞, 马蓝, 王旨意, 涂红霞, 杨绍凤, 王浩, 陈涛, 王小蓉. 中国樱桃杂交F1代花和果实若干性状遗传倾向分析[J]. 园艺学报, 2022, 49(9): 1853-1865. |
| [14] | 王沙, 张心慧, 赵玉洁, 李变变, 招雪晴, 沈雨, 董建梅, 苑兆和. 石榴花青苷合成相关基因PgMYB111的克隆与功能分析[J]. 园艺学报, 2022, 49(9): 1883-1894. |
| [15] | 黄玲, 胡先梅, 梁泽慧, 王艳平, 产祝龙, 向林. 郁金香花青素合成酶基因TgANS的克隆与功能鉴定[J]. 园艺学报, 2022, 49(9): 1935-1944. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司