
园艺学报 ›› 2022, Vol. 49 ›› Issue (1): 41-61.doi: 10.16420/j.issn.0513-353x.2020-0726
申序, 陈晓慧, 张婧, 陈荣珠, 徐小萍, 李晓斐, 蒋梦琦, 刘蒲东, 倪珊珊, 林玉玲, 赖钟雄( )
)
收稿日期:2021-09-15
修回日期:2021-12-17
出版日期:2022-01-25
发布日期:2022-01-24
通讯作者:
赖钟雄
E-mail:laizx01@163.com
基金资助:
SHEN Xu, CHEN Xiaohui, ZHANG Jing, CHEN Rongzhu, XU Xiaoping, LI Xiaofei, JIANG Mengqi, LIU Pudong, NI Shanshan, LIN Yuling, LAI Zhongxiong( )
)
Received:2021-09-15
Revised:2021-12-17
Online:2022-01-25
Published:2022-01-24
Contact:
LAI Zhongxiong
E-mail:laizx01@163.com
摘要:
以龙眼(Dimocarpus longan Lour.)‘红核子’品种早期体细胞胚胎发生的胚性愈伤组织(embryogenic callus,EC)、不完全胚性紧实结构(incomplete embryogenic compact structures,ICpEC)和球形胚(globular embryos,GE)为材料,通过生物信息学方法鉴定和分析其染色质重塑因子DlSnf2家族成员及分子进化特性,并通过转录组和实时荧光定量PCR(real-time quantitative PCR,qRT-PCR)分析其表达模式。龙眼DlSnf2家族包含35个成员,家族扩张受到全基因组或片段复制驱动。选择含有SNF2_N和HELICc结构域的31个成员,根据进化特征分为6大类19个亚族。蛋白互作、microRNA和启动子预测显示,部分成员能够发生互作,大部分基因受到microRNA的潜在调控,且存在与激素、胁迫、光、生长和发育相关顺式作用元件。转录组和qRT-PCR分析显示:大多数基因在ICpEC阶段中高表达,DlCHR18、DlCHR24、DlCHR25、DlCHR29和DlCHR31a从EC到GE的过程中表达逐渐下调,而DlCHR4、DlCHR15和DlCHR42表达逐渐上调。分析显示DlSnf2家族基因响应2,4-D、ABA、乙烯利、茉莉酸甲酯、4 ℃和40 ℃。DlCHR10和DlCHR11的表达被乙烯利上调,而DlCHR28b和DlCHR36被下调,DlCHR11被ABA和高温(40 ℃)下调,DlCHR10被茉莉酸甲酯下调,DlCHR36被低温(4 ℃)下调,DlCHR24和DlCHR35被高温(40 ℃)下调。Snf2基因家族在植物进化过程中可能具有功能的相对保守性及一定物种特异性。在植物生长调节剂和温度处理下,龙眼DlSnf2家族成员可能参与调控体胚发生和抵御高、低温胁迫,且推测DNA甲基化、microRNA和组蛋白修饰等其他表观调控机制参与了这一过程。
中图分类号:
申序, 陈晓慧, 张婧, 陈荣珠, 徐小萍, 李晓斐, 蒋梦琦, 刘蒲东, 倪珊珊, 林玉玲, 赖钟雄. 龙眼染色质重塑因子Snf2基因家族的进化动力学研究及在体胚发生早期的表达[J]. 园艺学报, 2022, 49(1): 41-61.
SHEN Xu, CHEN Xiaohui, ZHANG Jing, CHEN Rongzhu, XU Xiaoping, LI Xiaofei, JIANG Mengqi, LIU Pudong, NI Shanshan, LIN Yuling, LAI Zhongxiong. Evolutionary Dynamics Investigation and the Expression Analysis of Chromatin Remodeling Factor Snf2 Gene Family During Early Somatic Embryogenesis in Dimocarpus longan[J]. Acta Horticulturae Sinica, 2022, 49(1): 41-61.
| 基因 Gene | 正向引物(5′-3′) Forward primer | 反向引物(5′-3′) Reverse primer |
|---|---|---|
| DlCHR2 | AACTTCTCAGCATCGGGTGG | CCTCAAGTGGTGGTGGAACA |
| DlCHR6 | CCGGAGGTCTGGGAATCAAC | GGTCTGCCCAAGTCGATGAG |
| DlCHR8 | TTACGGGACTTGATCATGCC | TCTTTGTTCTGCAGTGAGGC |
| DlCHR10 | AAGTCGAGGGAGTTGCTTGG | GTCACGCTCAGAGGACACAA |
| DlCHR11 | TAAGGTACTCCGGCCGTTTC | AGGCGTTTACGTTCTCCACC |
| DlCHR12 | ACACCGTCTCGTCCATCTAT | AACCTGACATGCATTTTGGC |
| DlCHR14 | AGCTTCTTCCATTCCAGCGG | AGCCCAATGCAAACGTAAGG |
| DlCHR16b | GACCCTGATGATGATGTCCG | ACTGGTGGATGGGCTTAGAT |
| DlCHR21 | ACATTCATCCCACGAGCCAG | GCAGTAGCCTCTTGACCCAG |
| DlCHR24 | GCTGCGAGAACGTATTCAACC | GTGGTGAGCCATCAAAAGCC |
| DlCHR28b | GCATCTGCAATGATCCCCCT | GTGTGGCCTTGGAAAACACC |
| DlCHR29 | TGATACGCAACAGTACTCGG | TTTCGCAGCTCCTCTACAAC |
| DlCHR31a | GCTTGATGTTGCATGGAACC | GTCCCGGATGTAATGAGACG |
| DlCHR35 | GCTTTCGGGAACCCTCTACC | CGGTTCACAATAGGCCGAGA |
| DlCHR36 | GCAGCAGGCACATCAACAAT | TGGGTGCGTTGTATGCTCTT |
| DlCHR42 | AGTGCTCCGTGAAAGTCCTG | ATGAACTTCGGTCGAGCCAG |
| DlEF-1a | GATGATTCCCACCAAGCCCAT | GGGTCCTTCT TCTCAACACT CT |
| DleIF-4a | TTGTGCTGGATGAAGCTGATG | GGAAGGAGCTGGAAGATATCATAGA |
| DlFe-SOD | GGTCAGATGGTGAAGCCGTAGAG | GTCTATGCCACCGATACAACAAACCC |
表1 本研究中所用引物序列
Table 1 The primer sequences in this study
| 基因 Gene | 正向引物(5′-3′) Forward primer | 反向引物(5′-3′) Reverse primer |
|---|---|---|
| DlCHR2 | AACTTCTCAGCATCGGGTGG | CCTCAAGTGGTGGTGGAACA |
| DlCHR6 | CCGGAGGTCTGGGAATCAAC | GGTCTGCCCAAGTCGATGAG |
| DlCHR8 | TTACGGGACTTGATCATGCC | TCTTTGTTCTGCAGTGAGGC |
| DlCHR10 | AAGTCGAGGGAGTTGCTTGG | GTCACGCTCAGAGGACACAA |
| DlCHR11 | TAAGGTACTCCGGCCGTTTC | AGGCGTTTACGTTCTCCACC |
| DlCHR12 | ACACCGTCTCGTCCATCTAT | AACCTGACATGCATTTTGGC |
| DlCHR14 | AGCTTCTTCCATTCCAGCGG | AGCCCAATGCAAACGTAAGG |
| DlCHR16b | GACCCTGATGATGATGTCCG | ACTGGTGGATGGGCTTAGAT |
| DlCHR21 | ACATTCATCCCACGAGCCAG | GCAGTAGCCTCTTGACCCAG |
| DlCHR24 | GCTGCGAGAACGTATTCAACC | GTGGTGAGCCATCAAAAGCC |
| DlCHR28b | GCATCTGCAATGATCCCCCT | GTGTGGCCTTGGAAAACACC |
| DlCHR29 | TGATACGCAACAGTACTCGG | TTTCGCAGCTCCTCTACAAC |
| DlCHR31a | GCTTGATGTTGCATGGAACC | GTCCCGGATGTAATGAGACG |
| DlCHR35 | GCTTTCGGGAACCCTCTACC | CGGTTCACAATAGGCCGAGA |
| DlCHR36 | GCAGCAGGCACATCAACAAT | TGGGTGCGTTGTATGCTCTT |
| DlCHR42 | AGTGCTCCGTGAAAGTCCTG | ATGAACTTCGGTCGAGCCAG |
| DlEF-1a | GATGATTCCCACCAAGCCCAT | GGGTCCTTCT TCTCAACACT CT |
| DleIF-4a | TTGTGCTGGATGAAGCTGATG | GGAAGGAGCTGGAAGATATCATAGA |
| DlFe-SOD | GGTCAGATGGTGAAGCCGTAGAG | GTCTATGCCACCGATACAACAAACCC |
| 物种成员 Species member | 龙眼基因 Longan gene | Ka | Ks | Ka/Ks | 物种成员 Species member | 龙眼基因 Longan gene | Ka | Ks | Ka/Ks | |
|---|---|---|---|---|---|---|---|---|---|---|
| AtCHR33 | DlCHR28b | 0.40 | 1.36 | 0.30 | PNT24595 | DlCHR19 | 0.19 | 0.81 | 0.23 | |
| AtCHR30 | DlCHR28b | 0.50 | 1.45 | 0.35 | PNS97818 | DlCHR21 | 0.11 | 0.87 | 0.13 | |
| AtCHR15 | DlCHR15 | 0.73 | 1.85 | 0.39 | PNT29744 | DlCHR21 | 0.11 | 0.85 | 0.13 | |
| AtCHR9 | DlCHR9 | 0.23 | 2.85 | 0.08 | PNT11818 | DlCHR28b | 0.32 | 0.85 | 0.37 | |
| AtCHR37 | DlCHR37 | 0.27 | 1.50 | 0.18 | PNT39429 | DlCHR28b | 0.25 | 0.87 | 0.28 | |
| AtCHR18 | DlCHR18 | 0.17 | 1.09 | 0.16 | PNT41192 | DlCHR25 | 0.13 | 0.80 | 0.16 | |
| AtCHR34 | DlCHR35 | 0.45 | — | — | PNT41496 | DlCHR35 | 0.20 | 1.51 | 0.13 | |
| AtCHR35 | DlCHR35 | 0.31 | 2.02 | 0.15 | PNT20954 | DlCHR35 | 0.23 | 1.32 | 0.18 | |
| AtCHR6 | DlCHR6 | 0.21 | 1.82 | 0.12 | PNT56587 | DlCHR5 | 0.12 | 1.03 | 0.11 | |
| AtCHR36 | DlCHR36 | 0.27 | 1.38 | 0.19 | PNT42020 | DlCHR29 | 0.19 | 0.85 | 0.23 | |
| AtCHR10 | DlCHR10 | 0.20 | 1.34 | 0.15 | PNT42442 | DlCHR15 | 1.16 | 2.27 | 0.51 | |
| AtCHR2 | DlCHR2 | 0.20 | 1.31 | 0.15 | PNT19611 | DlCHR5 | 0.12 | 0.96 | 0.12 | |
| AtCHR25 | DlCHR25 | 0.19 | 1.18 | 0.16 | PNT05067 | DlCHR37 | 0.22 | 0.92 | 0.24 | |
| AtCHR12 | DlCHR12 | 0.18 | 1.70 | 0.10 | PNS94713 | DlCHR9 | 0.14 | 1.04 | 0.14 | |
| AtCHR11 | DlCHR11 | 0.06 | 1.67 | 0.03 | PNT09833 | DlCHR14 | 0.24 | 1.15 | 0.21 | |
| AtCHR7 | DlCHR6 | 0.32 | 2.14 | 0.15 | ar_scaffold00002.143 | DlCHR6 | 1.12 | 2.87 | 0.39 | |
| AtCHR29 | DlCHR29 | 0.21 | 1.29 | 0.16 | ar_scaffold00009.110 | DlCHR11 | 1.11 | — | — | |
| AtCHR17 | DlCHR11 | 0.06 | 1.57 | 0.04 | ar_scaffold00025.389 | DlCHR12 | 0.20 | 2.16 | 0.09 | |
| AtCHR23 | DlCHR12 | 0.20 | 1.60 | 0.12 | ar_scaffold00045.118 | DlCHR18 | 0.24 | 1.80 | 0.13 | |
| AtCHR22 | DlCHR36 | 0.95 | 2.20 | 0.43 | ar_scaffold00057.182 | DlCHR11 | 0.08 | 1.91 | 0.04 | |
| AtCHR24 | DlCHR24 | 0.32 | 1.78 | 0.18 | ar_scaffold00077.105 | DlCHR11 | 1.04 | 3.04 | 0.34 | |
| AtCHR1 | DlCHR1a | 0.21 | 2.17 | 0.10 | ar_scaffold00009.153 | DlCHR2 | 0.28 | 1.91 | 0.15 | |
| Os02t0762800-00 | DlCHR25 | 0.24 | 2.35 | 0.10 | ar_scaffold00045.118 | DlCHR2 | 0.95 | 2.30 | 0.41 | |
| Os02t0527100-01 | DlCHR29 | 0.25 | 2.42 | 0.10 | ar_scaffold00045.118 | DlCHR36 | 1.00 | 2.18 | 0.46 | |
| Os02t0650800-00 | DlCHR31a | 0.56 | — | — | ar_scaffold00067.58 | DlCHR8 | 0.26 | — | — | |
| Os03t0101700-01 | DlCHR10 | 0.52 | 2.15 | 0.24 | ar_scaffold00067.58 | DlCHR42 | 1.23 | 3.10 | 0.40 | |
| Os03t0722400-01 | DlCHR1a | 0.25 | — | — | ar_scaffold00077.105 | DlCHR36 | 0.35 | 1.67 | 0.21 | |
| Os05t0150300-01 | DlCHR11 | 0.10 | 3.63 | 0.03 | ar_scaffold00077.105 | DlCHR8 | 1.02 | 3.20 | 0.32 | |
| Os06t0183800-01 | DlCHR6 | 0.67 | 2.42 | 0.28 | ar_scaffold00077.105 | DlCHR2 | 1.15 | 3.54 | 0.32 | |
| Os07t0598300-01 | DlCHR14 | 0.40 | 2.10 | 0.19 | ar_scaffold00111.13 | DlCHR10 | 0.68 | 3.82 | 0.18 | |
| Os07t0636200-01 | DlCHR18 | 0.23 | 1.93 | 0.12 | ar_scaffold00009.153 | DlCHR1a | 0.72 | — | — | |
| PNT15539 | DlCHR12 | 0.13 | 0.83 | 0.15 | ar_scaffold00077.105 | DlCHR1a | 0.78 | 4.74 | 0.16 | |
| PNT13987 | DlCHR18 | 0.12 | 0.75 | 0.16 | ar_scaffold00029.1 | DlCHR28a | 0.35 | 1.40 | 0.25 | |
| PNT14263 | DlCHR11 | 0.04 | 0.97 | 0.04 | ar_scaffold00053.139 | DlCHR21 | 0.22 | 2.15 | 0.10 | |
| PNS92200 | DlCHR6 | 0.12 | 0.91 | 0.13 | ar_scaffold00003.327 | DlCHR28b | 0.25 | 1.56 | 0.16 | |
| PNT33877 | DlCHR6 | 0.12 | 0.95 | 0.13 | ar_scaffold00009.266 | DlCHR4 | 1.17 | 3.88 | 0.30 | |
| PNT25862 | DlCHR11 | 0.04 | 0.85 | 0.05 | ar_scaffold00067.58 | DlCHR4 | 0.87 | 3.48 | 0.25 | |
| PNT24755 | DlCHR12 | 0.13 | 0.90 | 0.14 | ar_scaffold00077.105 | DlCHR28b | 1.09 | 2.01 | 0.54 | |
| PNT25862 | DlCHR12 | 0.64 | — | — | ar_scaffold00146.48 | DlCHR4 | 1.10 | 3.01 | 0.37 | |
| PNT06750 | DlCHR20a | 0.16 | 0.79 | 0.20 | ar_scaffold00039.164 | DlCHR35 | 0.89 | — | — | |
| PNT07119 | DlCHR16a | 0.12 | 0.82 | 0.14 | ar_scaffold00126.55 | DlCHR25 | 0.22 | 2.06 | 0.11 | |
| PNT07119 | DlCHR16b | 0.12 | 0.82 | 0.14 | ar_scaffold00040.267 | DlCHR29 | 0.26 | 2.39 | 0.11 | |
| PNS90011 | DlCHR20a | 0.16 | 0.85 | 0.19 | ar_scaffold00148.60 | DlCHR15 | 0.60 | 2.25 | 0.27 | |
| PNT17288 | DlCHR42 | 0.27 | 1.28 | 0.21 | ar_scaffold00009.110 | DlCHR39 | 1.02 | 3.00 | 0.34 | |
| PNT03745 | DlCHR2 | 0.11 | 0.75 | 0.15 | ar_scaffold00045.118 | DlCHR37 | 1.09 | 2.28 | 0.48 | |
| PNT03205 | DlCHR10 | 0.18 | 0.89 | 0.20 | ar_scaffold00077.105 | DlCHR9 | 0.98 | — | — | |
| PNS90743 | DlCHR36 | 0.52 | — | — | ar_scaffold00002.143 | DlCHR13 | 1.21 | — | — | |
| PNT49991 | DlCHR2 | 0.11 | 0.81 | 0.14 | ar_scaffold00009.153 | DlCHR13 | 0.93 | 2.75 | 0.34 | |
| PNT38168 | DlCHR8 | 0.15 | 0.78 | 0.19 | ar_scaffold00032.254 | DlCHR13 | 0.82 | — | — | |
| PNT23278 | DlCHR42 | 0.24 | 1.04 | 0.23 | ar_scaffold00032.254 | DlCHR13 | 0.82 | — | — | |
| PNT35260 | DlCHR24 | 0.21 | 0.97 | 0.21 | ar_scaffold00009.110 | DlCHR31a | 1.06 | — | — | |
| PNT28173 | DlCHR24 | 0.21 | 0.89 | 0.24 | ar_scaffold00045.118 | DlCHR14 | 0.83 | — | — | |
| PNT15682 | DlCHR19 | 0.13 | 0.94 | 0.14 | ar_scaffold00110.12 | DlCHR14 | 0.36 | 2.13 | 0.17 |
表2 龙眼与拟南芥(At)、水稻(Os)、杨树(PN)及无油樟(ar)Snf2共线成员Ka/Ks分析
Table 2 Ka/Ks analysis of Snf2 collinear members in longan and Arabidopsis thaliana(At),Oryza sativa(Os),Populus trichocarpa(Pt),Amborella trichopoda(ar)
| 物种成员 Species member | 龙眼基因 Longan gene | Ka | Ks | Ka/Ks | 物种成员 Species member | 龙眼基因 Longan gene | Ka | Ks | Ka/Ks | |
|---|---|---|---|---|---|---|---|---|---|---|
| AtCHR33 | DlCHR28b | 0.40 | 1.36 | 0.30 | PNT24595 | DlCHR19 | 0.19 | 0.81 | 0.23 | |
| AtCHR30 | DlCHR28b | 0.50 | 1.45 | 0.35 | PNS97818 | DlCHR21 | 0.11 | 0.87 | 0.13 | |
| AtCHR15 | DlCHR15 | 0.73 | 1.85 | 0.39 | PNT29744 | DlCHR21 | 0.11 | 0.85 | 0.13 | |
| AtCHR9 | DlCHR9 | 0.23 | 2.85 | 0.08 | PNT11818 | DlCHR28b | 0.32 | 0.85 | 0.37 | |
| AtCHR37 | DlCHR37 | 0.27 | 1.50 | 0.18 | PNT39429 | DlCHR28b | 0.25 | 0.87 | 0.28 | |
| AtCHR18 | DlCHR18 | 0.17 | 1.09 | 0.16 | PNT41192 | DlCHR25 | 0.13 | 0.80 | 0.16 | |
| AtCHR34 | DlCHR35 | 0.45 | — | — | PNT41496 | DlCHR35 | 0.20 | 1.51 | 0.13 | |
| AtCHR35 | DlCHR35 | 0.31 | 2.02 | 0.15 | PNT20954 | DlCHR35 | 0.23 | 1.32 | 0.18 | |
| AtCHR6 | DlCHR6 | 0.21 | 1.82 | 0.12 | PNT56587 | DlCHR5 | 0.12 | 1.03 | 0.11 | |
| AtCHR36 | DlCHR36 | 0.27 | 1.38 | 0.19 | PNT42020 | DlCHR29 | 0.19 | 0.85 | 0.23 | |
| AtCHR10 | DlCHR10 | 0.20 | 1.34 | 0.15 | PNT42442 | DlCHR15 | 1.16 | 2.27 | 0.51 | |
| AtCHR2 | DlCHR2 | 0.20 | 1.31 | 0.15 | PNT19611 | DlCHR5 | 0.12 | 0.96 | 0.12 | |
| AtCHR25 | DlCHR25 | 0.19 | 1.18 | 0.16 | PNT05067 | DlCHR37 | 0.22 | 0.92 | 0.24 | |
| AtCHR12 | DlCHR12 | 0.18 | 1.70 | 0.10 | PNS94713 | DlCHR9 | 0.14 | 1.04 | 0.14 | |
| AtCHR11 | DlCHR11 | 0.06 | 1.67 | 0.03 | PNT09833 | DlCHR14 | 0.24 | 1.15 | 0.21 | |
| AtCHR7 | DlCHR6 | 0.32 | 2.14 | 0.15 | ar_scaffold00002.143 | DlCHR6 | 1.12 | 2.87 | 0.39 | |
| AtCHR29 | DlCHR29 | 0.21 | 1.29 | 0.16 | ar_scaffold00009.110 | DlCHR11 | 1.11 | — | — | |
| AtCHR17 | DlCHR11 | 0.06 | 1.57 | 0.04 | ar_scaffold00025.389 | DlCHR12 | 0.20 | 2.16 | 0.09 | |
| AtCHR23 | DlCHR12 | 0.20 | 1.60 | 0.12 | ar_scaffold00045.118 | DlCHR18 | 0.24 | 1.80 | 0.13 | |
| AtCHR22 | DlCHR36 | 0.95 | 2.20 | 0.43 | ar_scaffold00057.182 | DlCHR11 | 0.08 | 1.91 | 0.04 | |
| AtCHR24 | DlCHR24 | 0.32 | 1.78 | 0.18 | ar_scaffold00077.105 | DlCHR11 | 1.04 | 3.04 | 0.34 | |
| AtCHR1 | DlCHR1a | 0.21 | 2.17 | 0.10 | ar_scaffold00009.153 | DlCHR2 | 0.28 | 1.91 | 0.15 | |
| Os02t0762800-00 | DlCHR25 | 0.24 | 2.35 | 0.10 | ar_scaffold00045.118 | DlCHR2 | 0.95 | 2.30 | 0.41 | |
| Os02t0527100-01 | DlCHR29 | 0.25 | 2.42 | 0.10 | ar_scaffold00045.118 | DlCHR36 | 1.00 | 2.18 | 0.46 | |
| Os02t0650800-00 | DlCHR31a | 0.56 | — | — | ar_scaffold00067.58 | DlCHR8 | 0.26 | — | — | |
| Os03t0101700-01 | DlCHR10 | 0.52 | 2.15 | 0.24 | ar_scaffold00067.58 | DlCHR42 | 1.23 | 3.10 | 0.40 | |
| Os03t0722400-01 | DlCHR1a | 0.25 | — | — | ar_scaffold00077.105 | DlCHR36 | 0.35 | 1.67 | 0.21 | |
| Os05t0150300-01 | DlCHR11 | 0.10 | 3.63 | 0.03 | ar_scaffold00077.105 | DlCHR8 | 1.02 | 3.20 | 0.32 | |
| Os06t0183800-01 | DlCHR6 | 0.67 | 2.42 | 0.28 | ar_scaffold00077.105 | DlCHR2 | 1.15 | 3.54 | 0.32 | |
| Os07t0598300-01 | DlCHR14 | 0.40 | 2.10 | 0.19 | ar_scaffold00111.13 | DlCHR10 | 0.68 | 3.82 | 0.18 | |
| Os07t0636200-01 | DlCHR18 | 0.23 | 1.93 | 0.12 | ar_scaffold00009.153 | DlCHR1a | 0.72 | — | — | |
| PNT15539 | DlCHR12 | 0.13 | 0.83 | 0.15 | ar_scaffold00077.105 | DlCHR1a | 0.78 | 4.74 | 0.16 | |
| PNT13987 | DlCHR18 | 0.12 | 0.75 | 0.16 | ar_scaffold00029.1 | DlCHR28a | 0.35 | 1.40 | 0.25 | |
| PNT14263 | DlCHR11 | 0.04 | 0.97 | 0.04 | ar_scaffold00053.139 | DlCHR21 | 0.22 | 2.15 | 0.10 | |
| PNS92200 | DlCHR6 | 0.12 | 0.91 | 0.13 | ar_scaffold00003.327 | DlCHR28b | 0.25 | 1.56 | 0.16 | |
| PNT33877 | DlCHR6 | 0.12 | 0.95 | 0.13 | ar_scaffold00009.266 | DlCHR4 | 1.17 | 3.88 | 0.30 | |
| PNT25862 | DlCHR11 | 0.04 | 0.85 | 0.05 | ar_scaffold00067.58 | DlCHR4 | 0.87 | 3.48 | 0.25 | |
| PNT24755 | DlCHR12 | 0.13 | 0.90 | 0.14 | ar_scaffold00077.105 | DlCHR28b | 1.09 | 2.01 | 0.54 | |
| PNT25862 | DlCHR12 | 0.64 | — | — | ar_scaffold00146.48 | DlCHR4 | 1.10 | 3.01 | 0.37 | |
| PNT06750 | DlCHR20a | 0.16 | 0.79 | 0.20 | ar_scaffold00039.164 | DlCHR35 | 0.89 | — | — | |
| PNT07119 | DlCHR16a | 0.12 | 0.82 | 0.14 | ar_scaffold00126.55 | DlCHR25 | 0.22 | 2.06 | 0.11 | |
| PNT07119 | DlCHR16b | 0.12 | 0.82 | 0.14 | ar_scaffold00040.267 | DlCHR29 | 0.26 | 2.39 | 0.11 | |
| PNS90011 | DlCHR20a | 0.16 | 0.85 | 0.19 | ar_scaffold00148.60 | DlCHR15 | 0.60 | 2.25 | 0.27 | |
| PNT17288 | DlCHR42 | 0.27 | 1.28 | 0.21 | ar_scaffold00009.110 | DlCHR39 | 1.02 | 3.00 | 0.34 | |
| PNT03745 | DlCHR2 | 0.11 | 0.75 | 0.15 | ar_scaffold00045.118 | DlCHR37 | 1.09 | 2.28 | 0.48 | |
| PNT03205 | DlCHR10 | 0.18 | 0.89 | 0.20 | ar_scaffold00077.105 | DlCHR9 | 0.98 | — | — | |
| PNS90743 | DlCHR36 | 0.52 | — | — | ar_scaffold00002.143 | DlCHR13 | 1.21 | — | — | |
| PNT49991 | DlCHR2 | 0.11 | 0.81 | 0.14 | ar_scaffold00009.153 | DlCHR13 | 0.93 | 2.75 | 0.34 | |
| PNT38168 | DlCHR8 | 0.15 | 0.78 | 0.19 | ar_scaffold00032.254 | DlCHR13 | 0.82 | — | — | |
| PNT23278 | DlCHR42 | 0.24 | 1.04 | 0.23 | ar_scaffold00032.254 | DlCHR13 | 0.82 | — | — | |
| PNT35260 | DlCHR24 | 0.21 | 0.97 | 0.21 | ar_scaffold00009.110 | DlCHR31a | 1.06 | — | — | |
| PNT28173 | DlCHR24 | 0.21 | 0.89 | 0.24 | ar_scaffold00045.118 | DlCHR14 | 0.83 | — | — | |
| PNT15682 | DlCHR19 | 0.13 | 0.94 | 0.14 | ar_scaffold00110.12 | DlCHR14 | 0.36 | 2.13 | 0.17 |

图3 龙眼和7个物种Snf2蛋白的系统进化树分析 Dl:龙眼;At:拟南芥;Os:水稻;Pt:杨树;Ar:无油樟;Pa:挪威云杉;Pp:小立碗藓;Sm:江南卷柏。
Fig. 3 Phylogenetic tree analysis of Snf2 proteins from longan and other seven plant species Dl:Dimocarpus longan;At:Arabidopsis thaliana;Os:Oryza sativa;Pt:Populus trichocarpa;Ar:Amborella trichopoda;Pa:Picea abies;Pp:Physcomitrella patens;Sm:Selaginella moellendorffii.
| 基因 | 微小RNA | 期望值 | 抑制方式 | 基因 | 微小RNA | 期望值 | 抑制方式 | 基因 | 微小RNA | 期望值 | 抑制方式 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | MicroRNA | E-value | Inhibition | Gene | MicroRNA | E-value | Inhibition | Gene | MicroRNA | E-value | Inhibition | ||
| DlCHR1a | miR5177 | 3 | Cleavage | DlCHR12 | miR1312 | 3 | Cleavage | miR1514b | 4 | Cleavage | |||
| miR166j | 4 | Cleavage | miR1428e | 4 | Translation | miR3452 | 4 | Cleavage | |||||
| miR5079 | 4 | Cleavage | miR2868 | 4 | Cleavage | miR3637 | 4 | Cleavage | |||||
| DlCHR2 | miR2657a | 3 | Cleavage | miR396a | 4 | Cleavage | miR415 | 4 | Cleavage | ||||
| miR420 | 4 | Translation | DlCHR13 | miR414 | 3.5 | Cleavage | miR5078 | 4 | Translation | ||||
| miR4415 | 4 | Cleavage | miR416 | 3.5 | Cleavage | DlCHR24 | miR778 | 3.5 | Cleavage | ||||
| miR845d | 4 | Translation | miR847 | 3.5 | Cleavage | miR778 | 3.5 | Cleavage | |||||
| DlCHR3 | miR5200 | 3 | Cleavage | miR850 | 3.5 | Cleavage | miR414 | 4 | Cleavage | ||||
| miR1881 | 3.5 | Cleavage | miR860 | 3.5 | Cleavage | DlCHR25 | miR2936 | 3.5 | Cleavage | ||||
| miR1027a | 4 | Cleavage | miR869 | 3.5 | Cleavage | miR3631a | 4 | Cleavage | |||||
| miR1044 | 4 | Cleavage | miR1039 | 4 | Translation | DlCHR28a | miR1528 | 4 | Cleavage | ||||
| miR1521 | 4 | Cleavage | miR1044 | 4 | Cleavage | miR4249 | 4 | Cleavage | |||||
| miR166c | 4 | Cleavage | DlCHR14 | miR4400 | 2.5 | Cleavage | miR4366 | 4 | Cleavage | ||||
| miR2089*-3 | 4 | Cleavage | miR1535 | 4 | Cleavage | miR857 | 4 | Cleavage | |||||
| miR3707 | 4 | Cleavage | DlCHR15 | miR1160 | 3 | Cleavage | miR860 | 4 | Cleavage | ||||
| DlCHR4 | miR1884b | 3.5 | Cleavage | miR1528 | 4 | Translation | DlCHR28b | miR172a | 3.5 | Cleavage | |||
| miR2595 | 4 | Cleavage | miR1884b | 4 | Cleavage | miR1863 | 3.5 | Translation | |||||
| miR2615a | 4 | Translation | miR2611 | 4 | Cleavage | miR419 | 3.5 | Cleavage | |||||
| miR2616 | 4 | Translation | miR4223 | 4 | Cleavage | miR833 | 3.5 | Cleavage | |||||
| miR416 | 4 | Cleavage | DlCHR16a | miR1863 | 3 | Cleavage | DlCHR29 | miR3702 | 4 | Cleavage | |||
| miR860 | 4 | Cleavage | miR1863b | 3.5 | Cleavage | DlCHR31a | miR2673a | 2.5 | Cleavage | ||||
| DlCHR5 | miR2673a | 2.5 | Cleavage | miR1510a | 4 | Cleavage | miR2118e | 3 | Cleavage | ||||
| miR2673a | 4 | Translation | miR1871 | 4 | Cleavage | miR4249 | 3.5 | Cleavage | |||||
| DlCHR6 | miR2677 | 3.5 | Cleavage | miR2930 | 4 | Cleavage | miR1052 | 4 | Cleavage | ||||
| miR773b | 3.5 | Cleavage | miR3637 | 4 | Translation | miR2868 | 4 | Cleavage | |||||
| miR773b | 3.5 | Cleavage | miR5016 | 4 | Cleavage | DlCHR35 | miR870 | 3 | Translation | ||||
| miR1884b | 4 | Cleavage | miR820a | 4 | Cleavage | miR156e | 4 | Cleavage | |||||
| miR2105 | 4 | Cleavage | DlCHR16b | miR1863 | 3 | Cleavage | miR4391 | 4 | Cleavage | ||||
| miR2642 | 4 | Cleavage | miR1525 | 3.5 | Cleavage | miR5072 | 4 | Cleavage | |||||
| miR2928 | 4 | Cleavage | miR1863b | 3.5 | Cleavage | DlCHR36 | miR1028c | 3 | Cleavage | ||||
| miR870 | 4 | Cleavage | miR1510a | 4 | Cleavage | miR158b | 3.5 | Cleavage | |||||
| DlCHR8 | miR1863 | 4 | Cleavage | miR1871 | 4 | Cleavage | miR417 | 3.5 | Cleavage | ||||
| miR2928 | 4 | Cleavage | miR2930 | 4 | Cleavage | miR1081 | 4 | Translation | |||||
| miR4393b | 4 | Cleavage | miR3637 | 4 | Translation | miR1217 | 4 | Cleavage | |||||
| miR5021 | 4 | Cleavage | miR5016 | 4 | Cleavage | miR1919a | 4 | Cleavage | |||||
| miR5200 | 4 | Cleavage | miR820a | 4 | Cleavage | miR5052 | 4 | Cleavage | |||||
| DlCHR9 | miR1028c | 2.5 | Cleavage | DlCHR18 | miR390a.3 | 3.5 | Cleavage | miR5176 | 4 | Cleavage | |||
| miR3637 | 3.5 | Cleavage | miR536 | 3.5 | Cleavage | DlCHR37 | miR1027a | 4 | Cleavage | ||||
| miR393 | 3.5 | Translation | miR536 | 3.5 | Cleavage | miR1878 | 4 | Cleavage | |||||
| miR1320 | 4 | Cleavage | DlCHR19 | miR5025 | 3.5 | Cleavage | DlCHR39 | miR1169 | 3 | Cleavage | |||
| miR2604 | 4 | Cleavage | miR2097 | 4 | Translation | miR1888 | 4 | Cleavage | |||||
| miR860 | 4 | Cleavage | DlCHR20a | miR479 | 3 | Cleavage | miR2673a | 4 | Cleavage | ||||
| DlCHR11 | miR5058 | 3.5 | Cleavage | miR1513 | 4 | Cleavage | miR3706 | 4 | Cleavage | ||||
| miR1120 | 4 | Translation | miR2604 | 4 | Cleavage | DlCHR42 | miR1875 | 4 | Cleavage | ||||
| miR2610a | 4 | Cleavage | miR413 | 4 | Cleavage | miR2673a | 4 | Cleavage | |||||
| miR2673a | 4 | Cleavage | miR811d | 4 | Cleavage | miR3437 | 4 | Cleavage | |||||
| miR3435 | 4 | Cleavage | DlCHR21 | miR5079 | 3.5 | Cleavage | miR413 | 4 | Translation | ||||
| miR4246 | 4 | Cleavage | miR1044 | 4 | Translation | ||||||||
表3 调控龙眼DlSnf2家族基因的潜在microRNA分析
Table 3 Analysis of potential microRNA regulating longan DlSnf2 family genes
| 基因 | 微小RNA | 期望值 | 抑制方式 | 基因 | 微小RNA | 期望值 | 抑制方式 | 基因 | 微小RNA | 期望值 | 抑制方式 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | MicroRNA | E-value | Inhibition | Gene | MicroRNA | E-value | Inhibition | Gene | MicroRNA | E-value | Inhibition | ||
| DlCHR1a | miR5177 | 3 | Cleavage | DlCHR12 | miR1312 | 3 | Cleavage | miR1514b | 4 | Cleavage | |||
| miR166j | 4 | Cleavage | miR1428e | 4 | Translation | miR3452 | 4 | Cleavage | |||||
| miR5079 | 4 | Cleavage | miR2868 | 4 | Cleavage | miR3637 | 4 | Cleavage | |||||
| DlCHR2 | miR2657a | 3 | Cleavage | miR396a | 4 | Cleavage | miR415 | 4 | Cleavage | ||||
| miR420 | 4 | Translation | DlCHR13 | miR414 | 3.5 | Cleavage | miR5078 | 4 | Translation | ||||
| miR4415 | 4 | Cleavage | miR416 | 3.5 | Cleavage | DlCHR24 | miR778 | 3.5 | Cleavage | ||||
| miR845d | 4 | Translation | miR847 | 3.5 | Cleavage | miR778 | 3.5 | Cleavage | |||||
| DlCHR3 | miR5200 | 3 | Cleavage | miR850 | 3.5 | Cleavage | miR414 | 4 | Cleavage | ||||
| miR1881 | 3.5 | Cleavage | miR860 | 3.5 | Cleavage | DlCHR25 | miR2936 | 3.5 | Cleavage | ||||
| miR1027a | 4 | Cleavage | miR869 | 3.5 | Cleavage | miR3631a | 4 | Cleavage | |||||
| miR1044 | 4 | Cleavage | miR1039 | 4 | Translation | DlCHR28a | miR1528 | 4 | Cleavage | ||||
| miR1521 | 4 | Cleavage | miR1044 | 4 | Cleavage | miR4249 | 4 | Cleavage | |||||
| miR166c | 4 | Cleavage | DlCHR14 | miR4400 | 2.5 | Cleavage | miR4366 | 4 | Cleavage | ||||
| miR2089*-3 | 4 | Cleavage | miR1535 | 4 | Cleavage | miR857 | 4 | Cleavage | |||||
| miR3707 | 4 | Cleavage | DlCHR15 | miR1160 | 3 | Cleavage | miR860 | 4 | Cleavage | ||||
| DlCHR4 | miR1884b | 3.5 | Cleavage | miR1528 | 4 | Translation | DlCHR28b | miR172a | 3.5 | Cleavage | |||
| miR2595 | 4 | Cleavage | miR1884b | 4 | Cleavage | miR1863 | 3.5 | Translation | |||||
| miR2615a | 4 | Translation | miR2611 | 4 | Cleavage | miR419 | 3.5 | Cleavage | |||||
| miR2616 | 4 | Translation | miR4223 | 4 | Cleavage | miR833 | 3.5 | Cleavage | |||||
| miR416 | 4 | Cleavage | DlCHR16a | miR1863 | 3 | Cleavage | DlCHR29 | miR3702 | 4 | Cleavage | |||
| miR860 | 4 | Cleavage | miR1863b | 3.5 | Cleavage | DlCHR31a | miR2673a | 2.5 | Cleavage | ||||
| DlCHR5 | miR2673a | 2.5 | Cleavage | miR1510a | 4 | Cleavage | miR2118e | 3 | Cleavage | ||||
| miR2673a | 4 | Translation | miR1871 | 4 | Cleavage | miR4249 | 3.5 | Cleavage | |||||
| DlCHR6 | miR2677 | 3.5 | Cleavage | miR2930 | 4 | Cleavage | miR1052 | 4 | Cleavage | ||||
| miR773b | 3.5 | Cleavage | miR3637 | 4 | Translation | miR2868 | 4 | Cleavage | |||||
| miR773b | 3.5 | Cleavage | miR5016 | 4 | Cleavage | DlCHR35 | miR870 | 3 | Translation | ||||
| miR1884b | 4 | Cleavage | miR820a | 4 | Cleavage | miR156e | 4 | Cleavage | |||||
| miR2105 | 4 | Cleavage | DlCHR16b | miR1863 | 3 | Cleavage | miR4391 | 4 | Cleavage | ||||
| miR2642 | 4 | Cleavage | miR1525 | 3.5 | Cleavage | miR5072 | 4 | Cleavage | |||||
| miR2928 | 4 | Cleavage | miR1863b | 3.5 | Cleavage | DlCHR36 | miR1028c | 3 | Cleavage | ||||
| miR870 | 4 | Cleavage | miR1510a | 4 | Cleavage | miR158b | 3.5 | Cleavage | |||||
| DlCHR8 | miR1863 | 4 | Cleavage | miR1871 | 4 | Cleavage | miR417 | 3.5 | Cleavage | ||||
| miR2928 | 4 | Cleavage | miR2930 | 4 | Cleavage | miR1081 | 4 | Translation | |||||
| miR4393b | 4 | Cleavage | miR3637 | 4 | Translation | miR1217 | 4 | Cleavage | |||||
| miR5021 | 4 | Cleavage | miR5016 | 4 | Cleavage | miR1919a | 4 | Cleavage | |||||
| miR5200 | 4 | Cleavage | miR820a | 4 | Cleavage | miR5052 | 4 | Cleavage | |||||
| DlCHR9 | miR1028c | 2.5 | Cleavage | DlCHR18 | miR390a.3 | 3.5 | Cleavage | miR5176 | 4 | Cleavage | |||
| miR3637 | 3.5 | Cleavage | miR536 | 3.5 | Cleavage | DlCHR37 | miR1027a | 4 | Cleavage | ||||
| miR393 | 3.5 | Translation | miR536 | 3.5 | Cleavage | miR1878 | 4 | Cleavage | |||||
| miR1320 | 4 | Cleavage | DlCHR19 | miR5025 | 3.5 | Cleavage | DlCHR39 | miR1169 | 3 | Cleavage | |||
| miR2604 | 4 | Cleavage | miR2097 | 4 | Translation | miR1888 | 4 | Cleavage | |||||
| miR860 | 4 | Cleavage | DlCHR20a | miR479 | 3 | Cleavage | miR2673a | 4 | Cleavage | ||||
| DlCHR11 | miR5058 | 3.5 | Cleavage | miR1513 | 4 | Cleavage | miR3706 | 4 | Cleavage | ||||
| miR1120 | 4 | Translation | miR2604 | 4 | Cleavage | DlCHR42 | miR1875 | 4 | Cleavage | ||||
| miR2610a | 4 | Cleavage | miR413 | 4 | Cleavage | miR2673a | 4 | Cleavage | |||||
| miR2673a | 4 | Cleavage | miR811d | 4 | Cleavage | miR3437 | 4 | Cleavage | |||||
| miR3435 | 4 | Cleavage | DlCHR21 | miR5079 | 3.5 | Cleavage | miR413 | 4 | Translation | ||||
| miR4246 | 4 | Cleavage | miR1044 | 4 | Translation | ||||||||
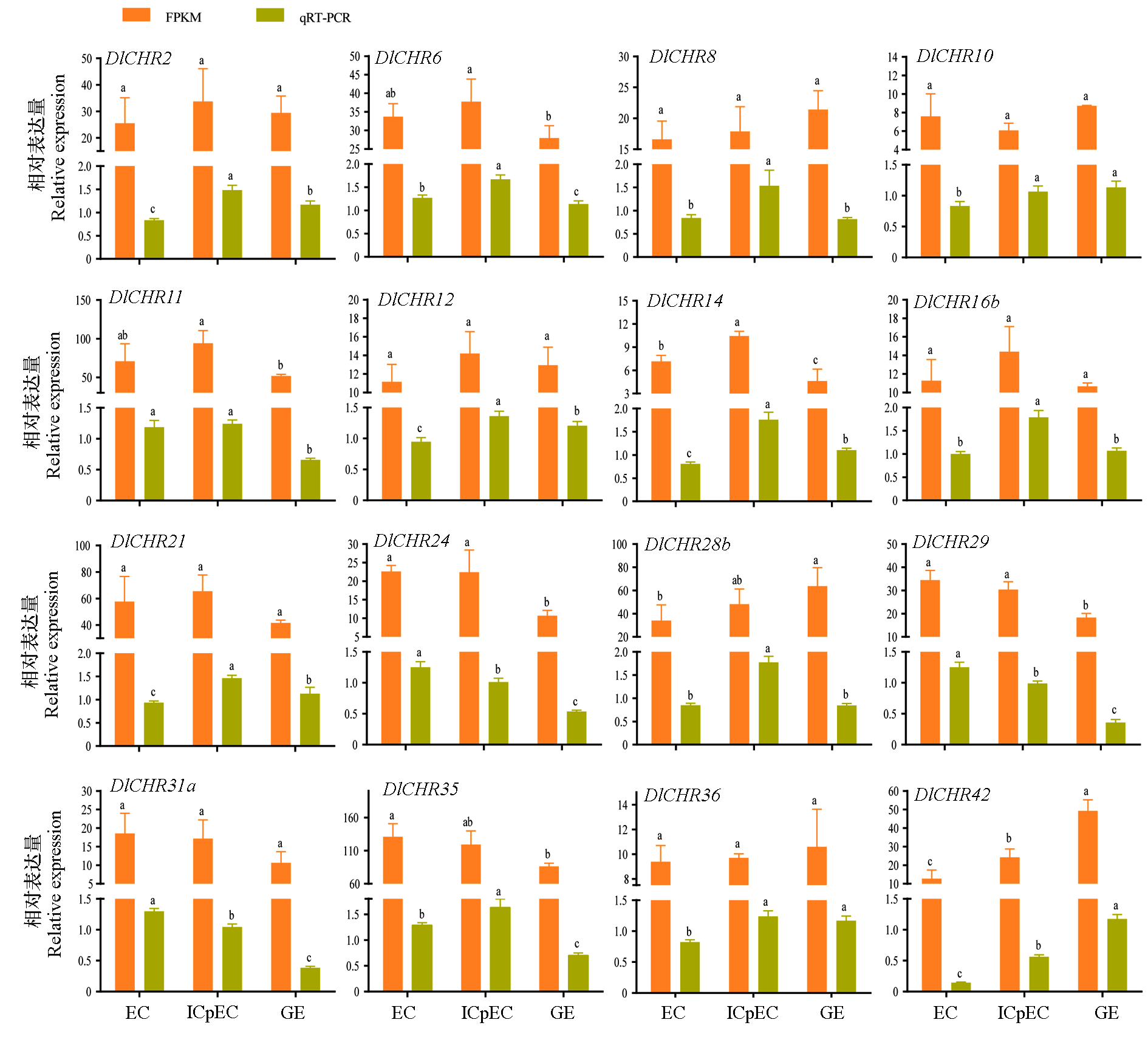
图6 龙眼体胚发生早期中DlSnf2成员的FPKM和qRT-PCR比较分析 数据被DlEF-1a、DlFe-SOD和DleIF-4a标准化。不同小写字母表示差异显著(P < 0.05,n = 3)。
Fig. 6 A comparative analysis of DlSnf2 members by FPKM and qRT-PCR in longan early somatic embryogenesis Data were normalized by DlEF-1a,DlFe-SOD and DleIF-4a.Different lowercase letters indicate significant difference(P < 0.05,n = 3).
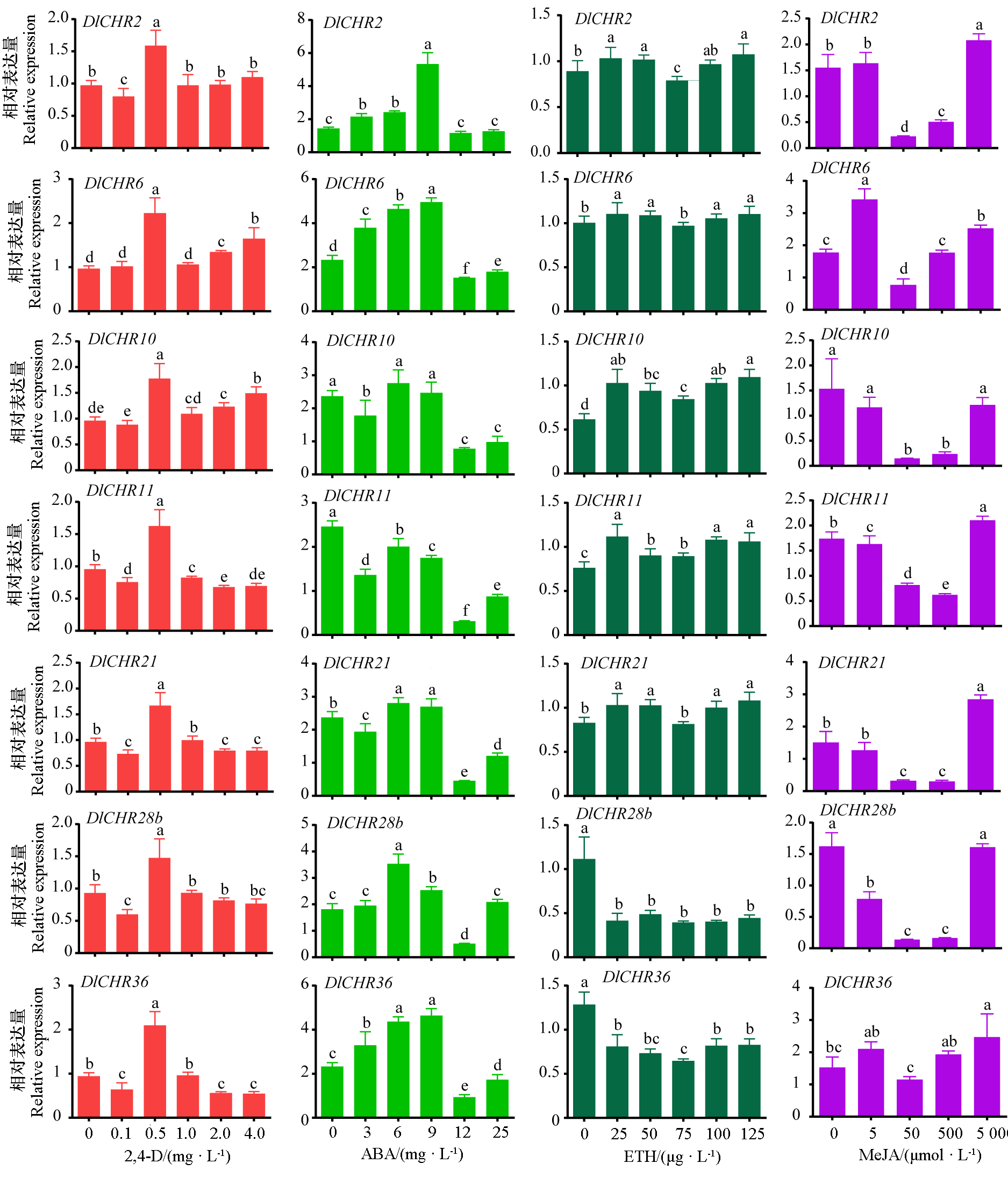
图7 不同植物生长调节剂处理下EC中DlSnf2成员表达分析 2,4-D组采用DlEF-1a和DleIF-4a标准化,其余组采用DlEF-1a、DlFe-SOD和DleIF-4a标准化。以不添加植物生长调节剂为对照,不同小写字母表示差异显著(P < 0.05,n = 3)。
Fig. 7 Expression analysis of DlSnf2 members in EC under different plant growth regulator treatments The 2,4-D group was normalized by DlEF-1a and DleIF-4a,other groups were normalized by DlEF-1a,DlFe-SOD and DleIF-4a. Samples without plant growth regulators were used as controls,and different lowercase letters indicate significant difference(P < 0.05,n = 3).
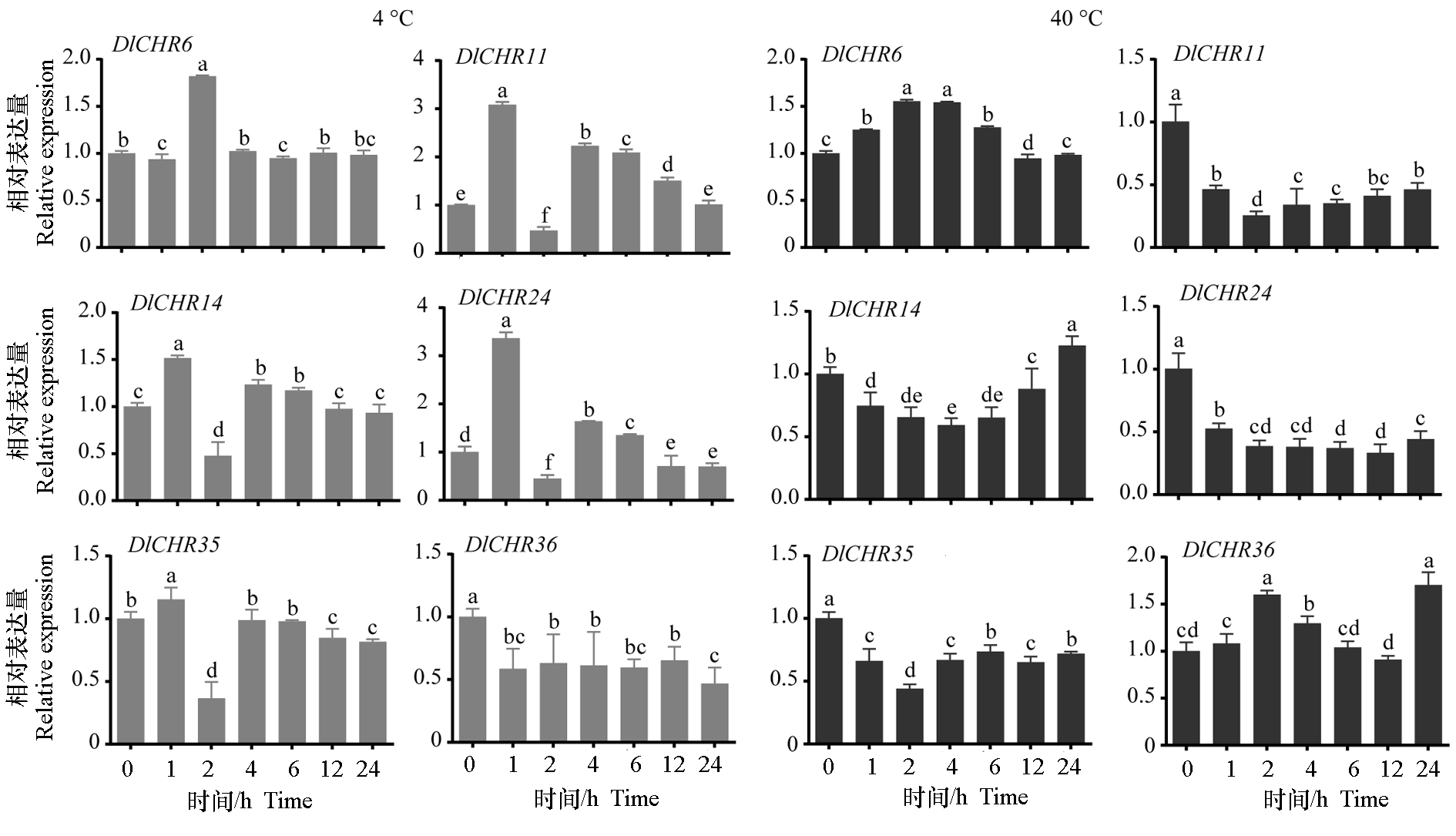
图8 高、低温处理下EC中DlSnf2成员的表达 数据被DleIF-4a标准化。不同小写字母表示差异显著(P < 0.05,n = 3)。
Fig. 8 Expression analysis of DlSnf2 members in EC under different temperature treatments Data were normalized by DleIF-4a. Different lowercase letters indicate significant difference(P < 0.05,n = 3).
| [1] |
Bargsten J W, Folta A, Mlynárová L, Nap J P. 2013. Snf 2 family gene distribution in higher plant genomes reveals DRD1 expansion and diversification in the tomato genome. PLoS ONE, 8 (11):e81147.
doi: 10.1371/journal.pone.0081147 URL |
| [2] |
Bouazoune K, Mitterweger A, Langst G, Imhof A, Akhtar A, Becker P B, Brehm A. 2002. The dMi-2 chromodomains are DNA binding modules important for ATP-dependent nucleosome mobilization. The EMBO Journal, 21 (10):2430-2440.
doi: 10.1093/emboj/21.10.2430 URL |
| [3] |
Busnelli S, Tripodi F, Nicastro R, Cirulli C, Tedeschi G, Pagliarin R, Alberghina L, Coccetti P. 2013. Snf1/AMPK promotes SBF and MBF-dependent transcription in budding yeast. Biochimica et Biophysica Acta, 1833 (12):3254-3264.
doi: S0167-4889(13)00336-4 pmid: 24084603 |
| [4] |
Buszewicz D, Archacki R, Palusiński A, Kotliński M, Fogtman A, Iwanickanowicka R, Sosnowska K, Kuciński J, Pupel P, Olędzki J, Dadlez M, Misicka A, Jerzmanowski A, Koblowska M. 2016. HD2C histone deacetylase and a SWI/SNF chromatin remodelling complex interact and both are involved in mediating the heat stress response in Arabidopsis. Plant Cell Environ, 39 (10):2108-2122.
doi: 10.1111/pce.v39.10 URL |
| [5] |
Chanvivattana Y, Bishopp A, Schubert D, Stock C, Moon Y, Sung Z R, Goodrich J. 2004. Interaction of polycomb-group proteins controlling flowering in Arabidopsis. Development, 131 (21):5263-5276.
pmid: 15456723 |
| [6] |
Chen C J, Chen H, Zhang Y, Thomas H R, Frank M H, He Y H, Xia R. 2020a. TBtools:an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 13 (8):1194-1202.
doi: 10.1016/j.molp.2020.06.009 URL |
| [7] |
Chen W, Zhu Q L, Liu Y G, Zhang Q Y. 2017. Chromatin remodeling and plant immunity. Advances in Protein Chemistry and Structural Biology, 106:243-260.
doi: S1876-1623(16)30050-5 pmid: 28057214 |
| [8] | Chen X H, Xu X P, Shen X, Li H S, Zhu C, Chen R Z, Nigarish Munir, Zhang Z H, Chen Y K, Xuhan X, Lin Y L, Lai Z X. 2020b. Genome-wide investigation of DNA methylation dynamics reveals a critical role of DNA demethylation during the early somatic embryogenesis of Dimocarpus longan Lour. Tree Physiology,(12):12. |
| [9] |
Deng W K, Wang Y B, Liu Z X, Cheng H, Xue Y. 2014. HemI:a toolkit for illustrating heatmaps. PLoS ONE, 9 (11):e111988.
doi: 10.1371/journal.pone.0111988 URL |
| [10] |
Erdel F, Rippe K. 2011. Chromatin remodelling in mammalian cells by ISWI-type complexes-where,when and why? Febs Journal, 278 (19):3608-3618.
doi: 10.1111/j.1742-4658.2011.08282.x URL |
| [11] |
Farrona S, Hurtado L, Bowman J L, Reyes J C. 2004. The Arabidopsis thaliana SNF 2 homolog AtBRM controls shoot development and flowering. Development, 131 (20):4965-4975.
pmid: 15371304 |
| [12] |
Flaus A, Martin D M, Barton G J, Owenhughes T. 2006. Identification of multiple distinct Snf 2 subfamilies with conserved structural motifs. Nucleic Acids Research, 34 (10):2887-2905.
doi: 10.1093/nar/gkl295 URL |
| [13] |
Folta A, Severing E I, Krauskopf J, van de Geest H, Verver J, Nap J P, Mlynarova L. 2014. Over-expression of Arabidopsis AtCHR23 chromatin remodeling ATPase results in increased variability of growth and gene expression. BMC Plant Biology, 14 (1):76.
doi: 10.1186/1471-2229-14-76 URL |
| [14] |
Fukaki H, Taniguchi N, Tasaka M. 2006. PICKLE is required for SOLITARY-ROOT/IAA14-mediated repression of ARF7 and ARF19 activity during Arabidopsis lateral root initiation. Plant Journal, 48 (3):380-389.
doi: 10.1111/tpj.2006.48.issue-3 URL |
| [15] |
Gerhold C B, Gasser S M. 2014. INO80 and SWR complexes:relating structure to function in chromatin remodeling. Trends in Cell Biology, 24 (11):619-631.
doi: 10.1016/j.tcb.2014.06.004 URL |
| [16] |
Guo F, Liu C L, Xia H, Bi Y P, Zhao C Z, Zhao S Z, Hou L, Li F G, Wang X J. 2013. Induced expression of AtLEC1 and AtLEC2 differentially promotes somatic embryogenesis in transgenic tobacco plants. PLoS ONE, 8 (8):e71714.
doi: 10.1371/journal.pone.0071714 URL |
| [17] |
Hahn J, Thiele D J. 2004. Activation of the Saccharomyces cerevisiae heat shock transcription factor under glucose starvation conditions by Snf 1 protein kinase. Journal of Biological Chemistry, 279 (7):5169-5176.
doi: 10.1074/jbc.M311005200 URL |
| [18] |
Han S K, Sang Y, Rodrigues A, F B, Wu M, Rodriguez P L, Wagner D. 2012. The SWI2/SNF 2 chromatin remodeling ATPase BRAHMA represses abscisic acid responses in the absence of the stress stimulus in Arabidopsis. The Plant Cell, 24 (12):4892-4906.
doi: 10.1105/tpc.112.105114 URL |
| [19] |
Han S K, Wu M, Cui S J, Wagner D. 2015. Roles and activities of chromatin remodeling ATPases in plants. Plant Journal, 83 (1):62-77.
doi: 10.1111/tpj.2015.83.issue-1 URL |
| [20] |
Hara T, Katoh H, Ogawa D, Kagaya Y, Sato Y, Kitano H, Nagato Y, Ishikawa R, Ono A, Kinoshita T, Takeda S, Hattori T. 2015. Rice SNF 2 family helicase ENL1 is essential for syncytial endosperm development. Plant Journal, 81 (1):1-12.
doi: 10.1111/tpj.12705 URL |
| [21] |
Holub E B. 2001. The arms race is ancient history in Arabidopsis,the wildflower. Nature Reviews Genetics, 2 (7):516-527.
pmid: 11433358 |
| [22] | Hu Y F, Liu D N, Zhong X C, Zhang C J, Zhang Q F, Zhou D X. 2012. CHD 3 protein recognizes and regulates methylated histone H3 lysines 4 and 27 over a subset of targets in the rice genome. Proceedings of the National Academy of Sciences of the United States of America, 109 (15):5773-5778. |
| [23] |
Hu Y F, Zhu N, Wang X M, Yi Q P, Zhu D Y, Lai Y, Zhao Y. 2013. Analysis of rice Snf2 family proteins and their potential roles in epigenetic regulation. Plant Physiology and Biochemistry, 70:33-42.
doi: 10.1016/j.plaphy.2013.05.001 URL |
| [24] | Huancamamani W, Garciaaguilar M, Leonmartinez G, Grossniklaus U, Viellecalzada J. 2005. CHR11,a chromatin-remodeling factor essential for nuclear proliferation during female gametogenesis in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America, 102 (47):17231-17236. |
| [25] |
Hurtado L, Farrona S, Reyes J C. 2006. The putative SWI/SNF complex subunit BRAHMA activates flower homeotic genes in Arabidopsis thaliana. Plant Molecular Biology, 62 (1):291-304.
doi: 10.1007/s11103-006-9021-2 URL |
| [26] | Jerzmanowski A. 2007. SWI/SNF chromatin remodeling and linker histones in plants. Biochimica et Biophysica Acta, 1769 (5):330-345. |
| [27] |
Knizewski L, Ginalski K, Jerzmanowski A. 2008. Snf 2 proteins in plants:gene silencing and beyond. Trends in Plant Science, 13 (10):557-565.
doi: 10.1016/j.tplants.2008.08.004 pmid: 18786849 |
| [28] |
Kwon C S, Hibara K, Pfluger J, Bezhani S, Metha H, Aida M, Tasaka M, Wagner D. 2006. A role for chromatin remodeling in regulation of CUC gene expression in the Arabidopsis cotyledon boundary. Development, 133 (16):3223-3230.
doi: 10.1242/dev.02508 URL |
| [29] | Lai Zhong-xiong. 2003. Study of longan biotechnology. Fuzhou: Fujian Science and Technology Press. (in Chinese) |
| 赖钟雄. 2003. 龙眼生物技术研究. 福州: 福建科学技术出版社. | |
| [30] |
Li C L, Chen C, Gao L, Yang S G, Nguyen V, Shi X J, Siminovitch K A, Kohalmi S E, Huang S Z, Wu K Q, Chen X M, Cui Y H. 2015. The Arabidopsis SWI2/SNF2 chromatin remodeler BRAHMA regulates polycomb function during vegetative development and directly activates the flowering repressor gene SVP. PLoS Genetics, 11 (1):e1004944.
doi: 10.1371/journal.pgen.1004944 URL |
| [31] |
Li G, Zhang J W, Li J Q, Yang Z N, Huang H, Xu L. 2012. Imitation Switch chromatin remodeling factors and their interacting RINGLET proteins act together in controlling the plant vegetative phase in Arabidopsis. Plant Journal, 72 (2):261-270.
doi: 10.1111/j.1365-313X.2012.05074.x URL |
| [32] |
Li Z X, Zhang L F, Li W F, Qi L W, Han S Y. 2017. MIR166a affects the germination of somatic embryos in Larix leptolepis by modulating IAA biosynthesis and signaling genes. Journal of Plant Growth Regulation, 36 (4):889-896.
doi: 10.1007/s00344-017-9693-7 URL |
| [33] |
Lin Y L, Lai Z X. 2010. Reference gene selection for qPCR analysis during somatic embryogenesis in longan tree. Plant Science, 178 (4):359-365.
doi: 10.1016/j.plantsci.2010.02.005 URL |
| [34] | Lin Y L, Min J M, Lai R L, Wu Z Y, Chen Y K, Yu L L, Cheng C Z, Jin Y C, Tian Q L, Liu Q F, Liu W H, Zhang C G, Lin L X, Hu Y, Zhang D M, Thu M, Zhang Z H, Liu S C, Zhong C S, Fang X D, Wang J, Yang H M, Varshney R K, Yin Y, Lai Z X. 2017. Genome-wide sequencing of longan(Dimocarpus longan Lour.)provides insights into molecular basis of its polyphenol-rich characteristics. GigaScience, 6 (5):1-14. |
| [35] |
Lotan T, Ohto M, Yee K M, West M A L, Lo R, Kwong R W, Yamagishi K, Fischer R L, Goldberg R B, Harada J J. 1998. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell, 93 (7):1195-1205.
pmid: 9657152 |
| [36] |
Makarevich G, Leroy O, Akinci U, Schubert D, Clarenz O, Goodrich J, Grossniklaus U, Kohler C. 2006. Different polycomb group complexes regulate common target genes in Arabidopsis. EMBO Reports, 7 (9):947-952.
pmid: 16878125 |
| [37] | Mccartney R R, Garnarwortzel L, Chandrashekarappa D G, Schmidt M C. 2016. Activation and inhibition of Snf 1 kinase activity by phosphorylation within the activation loop. Biochimica et Biophysica Acta, 1864 (11):1518-1528. |
| [38] |
Mlynarova L, Nap J P, Bisseling T. 2007. The SWI/SNF chromatin-remodeling gene AtCHR12 mediates temporary growth arrest in Arabidopsis thaliana upon perceiving environmental stress. Plant Journal, 51 (5):874-885.
doi: 10.1111/j.1365-313X.2007.03185.x URL |
| [39] |
Mu J Y, Tan H L, Zheng Q, Fu F Y, Liang Y, Zhang J, Yang X H, Wang T, Chong K, Wang X J, Zuo J R. 2008. LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiology, 148 (2):1042-1054.
doi: 10.1104/pp.108.126342 URL |
| [40] |
Noh Y, Amasino R M. 2003. PIE1,an ISWI family gene,is required for FLC activation and floral repression in Arabidopsis. The Plant Cell, 15 (7):1671-1682.
doi: 10.1105/tpc.012161 URL |
| [41] |
Ogas J, Cheng J, Sung Z R, Somerville C. 1997. Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science, 277 (5322):91-94.
pmid: 9204906 |
| [42] | Ogas J, Kaufmann S H, Henderson J T, Somerville C. 1999. PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 96 (24):13839-13844. |
| [43] |
Pessina S, Tsiarentsyeva V, Busnelli S, Vanoni M, Alberghina L, Coccetti P. 2010. Snf1/AMPK promotes S-phase entrance by controlling CLB5 transcription in budding yeast. Cell Cycle, 9 (11):2189-2200.
doi: 10.4161/cc.9.11.11847 URL |
| [44] |
Reyes J C. 2014. The many faces of plant SWI/SNF complex. Molecular Plant, 7 (3):454-458.
doi: 10.1093/mp/sst147 pmid: 24177686 |
| [45] | Riemann M, Dhakarey R, Hazman M, Miro B, Kohli A, Nick P. 2015. Exploring jasmonates in the hormonal network of drought and salinity responses. Frontiers in Plant Science, 6:1077. |
| [46] |
Rizhsky L, Liang H, Shuman J L, Shulaev V, Davletova S, Mittler R. 2004. When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiology, 134 (4):1683-1696.
doi: 10.1104/pp.103.033431 URL |
| [47] | Sanchez R, Zhou M M. 2009. The role of human bromodomains in chromatin biology and gene transcription. Current Opinion in Drug Discovery & Development, 12 (5):659-665. |
| [48] |
Sang Y, Silvaortega C O, Wu S, Yamaguchi N, Wu M, Pfluger J. 2012. Mutations in two non-canonical Arabidopsis SWI2/SNF 2 chromatin remodeling ATPases cause embryogenesis and stem cell maintenance defects. Plant Journal, 72 (6):1000-1014.
doi: 10.1111/tpj.2012.72.issue-6 URL |
| [49] |
Sarnowski T J, Rios G, Jasik J, Świezewski S, Kaczanowski S, Li Y, Kwiatkowska A, Pawlikowska K, Koźbial M, Koźbial P, Koncz C, Jerzmanowski A. 2005. SWI 3 subunits of putative SWI/SNF chromatin-remodeling complexes play distinct roles during Arabidopsis development. The Plant Cell, 17 (9):2454-2472.
doi: 10.1105/tpc.105.031203 URL |
| [50] |
Schmutz J, Cannon S B, Schlueter J, Ma J X, Mitros T, Nelson W, Hyten D L, Song Q J, Thelen J J, Cheng J L, Xu D, Hellsten U, May G D, Yu Y, Sakurai T, Umezawa T, Bhattacharyya M K, Sandhu D, Valliyodan B, Lindquist E, Peto M, Grant D, Shu S Q, Goodstein D, Barry K, Futrell-Griggs M, Abernathy B, Du J C, Tian Z X, Zhu L C, Gill N, Joshi T, Libault M, Sethuraman A, Zhang X C, Shinozaki K, Nguyen H T, Wing R A, Cregan P, Specht J, Grimwood J, Rokhsar, Stacey G, Shoemaker R C, Jackson S A. 2010. Genome sequence of the palaeopolyploid soybean. Nature, 463 (7278):178-183.
doi: 10.1038/nature08670 URL |
| [51] |
Shaked H, Aviviragolsky N, Levy A A. 2006. Involvement of the Arabidopsis SWI2/SNF 2 chromatin remodeling gene family in DNA damage response and recombination. Genetics, 173 (2):985-994.
pmid: 16547115 |
| [52] | Shen Xu, Chen Xiao-hui, Xu Xiao-ping, Huo Wen, Li Xiao-fei, Jiang Meng-qi, Zhang Jing, Lin Yu-lin, Lai Zhong-xiong. 2019. Genome-wide identification and expression analysis of SDG gene family during early somatic embryogenesis in Dimocarpus longan Lour. Chinese Journal of Tropical Crops, 40 (10):1889-1901. (in Chinese) |
| 申序, 陈晓慧, 徐小萍, 霍雯, 李晓斐, 蒋梦琦, 张婧, 林玉玲, 赖钟雄. 2019. 龙眼体细胞胚胎发生早期SDG基因家族的全基因组鉴定与表达分析. 热带作物学报, 40 (10):1889-1901. | |
| [53] |
Shibukawa T, Yazawa K, Kikuchi A, Kamada H. 2009. Possible involvement of DNA methylation on expression regulation of carrot LEC1 gene in its 5′-upstream region. Gene, 437 (1-2):22-31.
doi: 10.1016/j.gene.2009.02.011 pmid: 19264116 |
| [54] | Simpsonlavy K J, Johnston M. 2013. SUMOylation regulates the SNF1 protein kinase. Proceedings of the National Academy of Sciences of the United States of America, 110 (43):17432-17437. |
| [55] | Smaczniak C, Immink R G, Muino J M, Blanvillain R, Busscher M, Busscherlange J, Dung-Dinh Q, Liu S J, Westphal A H, Boeren S, Parcy F, Xu L, Carles C C, Angenent G C, Kaufmann K. 2012. Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proceedings of the National Academy of Sciences of the United States of America, 109 (5):1560-1565. |
| [56] | Stone S L, Braybrook S A, Paula S L, Kwong L W, Meuser J E, Pelletier J M, Hsieh T, Fischer R L, Goldberg R B, Harada J J. 2008. Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity:implications for somatic embryogenesis. Proceedings of the National Academy of Sciences of the United States of America, 105 (8):3151-3156. |
| [57] |
Wagner D, Meyerowitz E M. 2002. SPLAYED,a novel SWI/SNF ATPase homolog,controls reproductive development in Arabidopsis. Current Biology, 12 (2):85-94.
pmid: 11818058 |
| [58] |
Walley J W, Rowe H C, Xiao Y M, Chehab E W, Kliebenstein D J, Wagner D, Dehesh K. 2008. The chromatin remodeler SPLAYED regulates specific stress signaling pathways. PLoS Pathogens, 4 (12):e1000237.
doi: 10.1371/journal.ppat.1000237 URL |
| [59] |
Wang Y P, Tang H B, Debarry J D, Tan X F, Li J P, Wang X Y, Lee T, Jin H Z, Marler B, Guo H, Kissinger J C, Paterson A H. 2012. MCScanX:a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Research, 40 (7):e49.
doi: 10.1093/nar/gkr1293 URL |
| [60] |
Wilson M A, Koutelou E, Hirsch C L, Akdemir K C, Schibler A C, Barton M C, Dent S Y. 2011. Ubp8 and SAGA regulate Snf 1 AMP kinase activity. Molecular and Cellular Biology, 31 (15):3126-3135.
doi: 10.1128/MCB.01350-10 URL |
| [61] |
Wójcik A M, Nodine M D, Gaj M D. 2017. miR160 and miR166/ 165 contribute to the LEC2-mediated auxin response involved in the somatic embryogenesis induction in Arabidopsis. Frontiers in Plant Science, 8:2024.
doi: 10.3389/fpls.2017.02024 pmid: 29321785 |
| [62] |
Wójcikowska B, Jaskóła K, Gąsiorek P, Meus M, Nowak K, Gaj M D. 2013. LEAFY COTYLEDON2(LEC2)promotes embryogenic induction in somatic tissues of Arabidopsis,via YUCCA-mediated auxin biosynthesis. Planta, 238 (3):425-440.
doi: 10.1007/s00425-013-1892-2 pmid: 23722561 |
| [63] |
Yang D L, Zhang G P, Wang L L, Li J W, Xu D C, Di C R, Tang K, Yang L, Zeng L, Miki D, Duan C G, Zhang H M, Zhu J K. 2018. Four putative SWI2/SNF 2 chromatin remodelers have dual roles in regulating DNA methylation in Arabidopsis. Cell Discovery, 4 (1):55.
doi: 10.1038/s41421-018-0056-8 URL |
| [64] |
Yang X Y, Wang L C, Yuan D J, Lindsey K, Zhang X L. 2013. Small RNA and degradome sequencing reveal complex miRNA regulation during cotton somatic embryogenesis. Journal of Experimental Botany, 64 (6):1521-1536.
doi: 10.1093/jxb/ert013 URL |
| [65] |
Zhang D D, Gao S J, Yang P, Yang J, Yang S G, Wu K Q. 2019. Identification and expression analysis of Snf 2 family proteins in tomato (Solanum lycopersicum). Comparative and Functional Genomics,doi: 10.1155/2019/5080935.
doi: 10.1155/2019/5080935 |
| [66] | Zhang Qinglin, Su Liyao, Li Xue, Zhang Shuting, Xu Xiaoping, Chen Xiaohui, Wang Peiyu, Li Rong, Zhang Zihao, Chen Yukun, Lai Zhongxiong, Lin Yuling. 2018. Cloning and expression analysis of miR166 primary gene during the early stage of somatic embryogenesis of longan. Acta Horticulturae Sinica, 45 (8):1501-1512. (in Chinese) |
| 张清林, 苏立遥, 厉雪, 张舒婷, 徐小萍, 陈晓慧, 王培育, 李蓉, 张梓浩, 陈裕坤, 赖钟雄, 林玉玲. 2018. 龙眼体胚发生miR166初级体的克隆与表达分析. 园艺学报, 45 (8):1501-1512. | |
| [67] |
Zhang S Z, Liu X G, Lin Y A, Xie G N, Fu F L, Liu H L, Wang J, Gao S B, Lan H, Rong T Z. 2011. Characterization of a ZmSERK gene and its relationship to somatic embryogenesis in a maize culture. Plant Cell Tissue and Organ Culture, 105 (1):29-37.
doi: 10.1007/s11240-010-9834-1 URL |
| [68] |
Zhao M L, Yang S G, Chen C, Li C L, Shan W, Lu W J, Cui Y H. 2015. Arabidopsis BREVIPEDICELLUS interacts with the SWI2/SNF 2 chromatin remodeling ATPase BRAHMA to regulate KNAT2 and KNAT6 expression in control of inflorescence architecture. PLoS Genetics, 11 (3):e1005125.
doi: 10.1371/journal.pgen.1005125 URL |
| [69] |
Zhu J K, Hasegawa P M, Bressan R A. 1997. Molecular aspects of osmotic stress in plants. Critical Reviews in Plant Sciences, 16 (3):253-277.
doi: 10.1080/07352689709701950 URL |
| [1] | 邓朝军, 许奇志, 蒋际谋, 胡文舜, 郑少泉, 陈秀萍, 姜 帆, 许家辉, 苏文炳, 张雅玲, 黄敬峰. 浓香型龙眼新品种‘醇香’[J]. 园艺学报, 2022, 49(S2): 75-76. |
| [2] | 邓朝军, 陈秀萍, 许奇志, 蒋际谋, 郑少泉, 胡文舜, 姜 帆, 许家辉, 苏文炳, 张雅玲, 黄敬峰. 浓香型龙眼新品种‘福香’[J]. 园艺学报, 2022, 49(S2): 77-78. |
| [3] | 徐小萍, 曹清影, 蔡柔荻, 官庆栩, 张梓浩, 陈裕坤, 徐涵, 林玉玲, 赖钟雄. 龙眼miR408与DlLAC12克隆及其在球形胚发生和非生物胁迫下的表达分析[J]. 园艺学报, 2022, 49(9): 1866-1882. |
| [4] | 邹学校, 朱凡. 辣椒的起源、进化与栽培历史[J]. 园艺学报, 2022, 49(6): 1371-1381. |
| [5] | 梁沁, 张延晖, 康开权, 刘瑾航, 李亮, 冯宇, 王超, 杨超, 李永裕. miR168家族进化特性及其在砂梨休眠期的表达模式分析[J]. 园艺学报, 2022, 49(5): 958-972. |
| [6] | 梁晨, 孙如意, 向锐, 孙艺萌, 师校欣, 杜国强, 王莉. 葡萄生长调控因子GRF家族基因的鉴定及表达分析[J]. 园艺学报, 2022, 49(5): 995-1007. |
| [7] | 肖学宸, 刘梦雨, 蒋梦琦, 陈燕, 薛晓东, 周承哲, 吴兴健, 吴君楠, 郭寅生, 叶开温, 赖钟雄, 林玉玲. 龙眼褪黑素合成途径SNAT、ASMT和COMT家族基因鉴定及表达分析[J]. 园艺学报, 2022, 49(5): 1031-1046. |
| [8] | 潘鑫峰, 叶方婷, 毛志君, 李兆伟, 范凯. 睡莲WRKY家族的全基因组鉴定和分子进化分析[J]. 园艺学报, 2022, 49(5): 1121-1135. |
| [9] | 刘梦雨, 蒋梦琦, 陈燕, 张舒婷, 薛晓东, 肖学宸, 赖钟雄, 林玉玲. 龙眼GDSL酯酶/脂肪酶基因的全基因组鉴定及表达分析[J]. 园艺学报, 2022, 49(3): 597-612. |
| [10] | 武晓晓, 陈传武, 刘萍, 唐艳, 邓崇岭. 基于重测序的广西姑婆山野生柑橘资源遗传演化及分类地位分析[J]. 园艺学报, 2022, 49(2): 407-415. |
| [11] | 冯丽肖, 胡荣, 卜姗, 张德咏, 罗香文, 李凡, 丁铭, 张卓, 张松柏, 刘勇. 云南省番茄莴苣褪绿病毒分子检测及遗传进化分析[J]. 园艺学报, 2022, 49(1): 141-147. |
| [12] | 张春渝, 许小琼, 徐小萍, 赵鹏程, 申序, MunirNigarish, 张梓浩, 林玉玲, 陈振光, 赖钟雄. 龙眼SKP1-like家族成员鉴定及体胚发生早期表达分析[J]. 园艺学报, 2021, 48(9): 1665-1679. |
| [13] | 杨为海, 曾利珍, 肖秋生, 石胜友. 饥饿胁迫下龙眼落果与果皮和离区糖、ABA及相关基因表达的变化[J]. 园艺学报, 2021, 48(8): 1457-1469. |
| [14] | 苏立遥, 王培育, 蒋梦琦, 黄倏祺, 薛晓东, 刘梦雨, 肖学宸, 赖春旺, 张梓浩, 陈裕坤, 赖钟雄, 林玉玲. 龙眼pri-miR319a编码短肽活性的研究[J]. 园艺学报, 2021, 48(5): 908-920. |
| [15] | 蔡柔荻, 厉雪, 陈燕, 徐小萍, 陈晓慧, 赖钟雄, 林玉玲. 龙眼DRB家族全基因组鉴定及其表达分析[J]. 园艺学报, 2021, 48(5): 921-933. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司