
园艺学报 ›› 2021, Vol. 48 ›› Issue (11): 2262-2274.doi: 10.16420/j.issn.0513-353x.2020-0944
庄玥莹, 周利君, 程璧瑄, 于超( ), 罗乐, 潘会堂, 张启翔
), 罗乐, 潘会堂, 张启翔
收稿日期:2021-04-09
修回日期:2021-08-06
发布日期:2021-12-02
基金资助:
ZHUANG Yueying, ZHOU Lijun, CHENG Bixuan, YU Chao( ), LUO Le, PAN Huitang, ZHANG Qixiang
), LUO Le, PAN Huitang, ZHANG Qixiang
Received:2021-04-09
Revised:2021-08-06
Published:2021-12-02
摘要:
通过转录组测序技术,探究大花香水月季及单瓣香水月季两个变种的花香代谢通路及花香差异形成的相关基因。在两个材料花蕾期、初开期及盛开期对比组中分别检测出8 339、8 849、7 572个差异表达基因,其中3个阶段共有的差异基因3 762个。GO和KEGG聚类分析显示,差异基因主要参与代谢过程、细胞过程、单一有机体过程、催化活性等生物学过程;主要富集于代谢途径、次生代谢物的生物合成、碳代谢等途径。在3组共有差异基因中共发现富集于6条重要花香物质合成通路(萜类骨架生物合成、苯丙素生物合成、脂肪酸生物合成、单萜类生物合成、双萜类生物合成及倍半萜和三萜类生物合成)中的差异基因27个。认为这27个基因对两个变种花香释放过程中的差异形成有较大影响。分析特征花香物质合成酶基因Rc_0128091及Rc_0119291在转录组数据中的表达,发现其表达量与相应的花香物质3,5-二甲氧基苯及1,3,5-三甲氧基苯释放量趋势基本一致,推测对特征花香物质的合成有决定性作用。随机选取10个差异表达基因进行qRT-PCR和RNA-seq的相关性验证,结果显示二者相关性较高,认为RNA-seq测序结果具有较高的准确性。
中图分类号:
庄玥莹, 周利君, 程璧瑄, 于超, 罗乐, 潘会堂, 张启翔. 基于转录组测序的香水月季花香代谢基因研究[J]. 园艺学报, 2021, 48(11): 2262-2274.
ZHUANG Yueying, ZHOU Lijun, CHENG Bixuan, YU Chao, LUO Le, PAN Huitang, ZHANG Qixiang. Study on the Fragrance Metabolic Genes of Rosa odorata Based on Transcriptome Sequencing[J]. Acta Horticulturae Sinica, 2021, 48(11): 2262-2274.

图1 本研究中的试验材料 T1:大花香水月季;T2:单瓣香水月季;a:花蕾期;b:初开期;c:全开期。
Fig. 1 Experimental materials for this study T1:Rosa odorata var. gigantea;T2:R. odorata var. odorata;a:Flower budding stage;b:Early flowering stage;c:Blooming stage.
| 基因ID Gene ID | 正向引物序列(5′-3′) Forward primer sequence | 反向引物序列(5′-3′) Reverse primer sequence |
|---|---|---|
| GAPDH | ATCCATTCATCACCACCGACTACA | GCATCCTTACTTGGGGCAGAGA |
| Rc 0174951 | TGGTTTCCCTAGGAGTCACG | GGAAGTGGTGCATAAGGGAT |
| Rc 0121071 | AGAAAGGCCATCCATAGTGC | TTGGCCTTCTTGATTGTCTC |
| Rc 0059511 | TGGATTTCAACCTGCTGCA | TCTCACGTCCAAGTCCTTCC |
| Rc 0412071 | TGAAACTTGATTCAGGCAAGC | TTCTGGGGTGGAGGACCTT |
| Rc 0241141 | ATGGCATTTAACACTGCTGGT | CGGCTCCTGCTTAAGAACCT |
| Rc 0083751 | TCAGTGGCTATGTTGGTGGT | ACCAATCAGCCTAAAATGCC |
| Rc 0392511 | CTCCTCCAGTCTGGCCAAAT | TGACATGTATGTTTTCCGCGT |
| Rc 0110711 | ACTCAACATTCCTGCAACGA | AGCGGACAGGTAGTTTTTGA |
| Rc 0214071 | CTGCTGGGGAAGAGAAGAGT | ATGGAGGACGAAACATTTGC |
表1 qRT-PCR引物序列
Table 1 qRT-PCR primers sequences
| 基因ID Gene ID | 正向引物序列(5′-3′) Forward primer sequence | 反向引物序列(5′-3′) Reverse primer sequence |
|---|---|---|
| GAPDH | ATCCATTCATCACCACCGACTACA | GCATCCTTACTTGGGGCAGAGA |
| Rc 0174951 | TGGTTTCCCTAGGAGTCACG | GGAAGTGGTGCATAAGGGAT |
| Rc 0121071 | AGAAAGGCCATCCATAGTGC | TTGGCCTTCTTGATTGTCTC |
| Rc 0059511 | TGGATTTCAACCTGCTGCA | TCTCACGTCCAAGTCCTTCC |
| Rc 0412071 | TGAAACTTGATTCAGGCAAGC | TTCTGGGGTGGAGGACCTT |
| Rc 0241141 | ATGGCATTTAACACTGCTGGT | CGGCTCCTGCTTAAGAACCT |
| Rc 0083751 | TCAGTGGCTATGTTGGTGGT | ACCAATCAGCCTAAAATGCC |
| Rc 0392511 | CTCCTCCAGTCTGGCCAAAT | TGACATGTATGTTTTCCGCGT |
| Rc 0110711 | ACTCAACATTCCTGCAACGA | AGCGGACAGGTAGTTTTTGA |
| Rc 0214071 | CTGCTGGGGAAGAGAAGAGT | ATGGAGGACGAAACATTTGC |
| 样本 Sample | 原始测序数据 Raw reads | 过滤后测序数据 Clean reads | 未比对测序数据 Unmapped reads | Q20/% | Q30/% | GC/% |
|---|---|---|---|---|---|---|
| T1a | 62 786 040 | 62 674 405 | 60 810 922 | 97.83 | 93.80 | 47.62 |
| T1b | 67 274 777 | 67 166 924 | 65 797 731 | 98.17 | 94.59 | 47.80 |
| T1c | 61 386 237 | 61 283 162 | 60 016 880 | 97.85 | 93.82 | 47.71 |
| T2a | 66 064 221 | 65 936 595 | 61 819 035 | 97.94 | 94.03 | 47.93 |
| T2b | 65 163 741 | 65 017 603 | 61 516 931 | 97.48 | 93.01 | 47.94 |
| T2c | 62 902 745 | 62 794 761 | 58 409 372 | 98.06 | 94.28 | 48.21 |
表2 转录组测序结果统计
Table 2 RNA-seq data statistics
| 样本 Sample | 原始测序数据 Raw reads | 过滤后测序数据 Clean reads | 未比对测序数据 Unmapped reads | Q20/% | Q30/% | GC/% |
|---|---|---|---|---|---|---|
| T1a | 62 786 040 | 62 674 405 | 60 810 922 | 97.83 | 93.80 | 47.62 |
| T1b | 67 274 777 | 67 166 924 | 65 797 731 | 98.17 | 94.59 | 47.80 |
| T1c | 61 386 237 | 61 283 162 | 60 016 880 | 97.85 | 93.82 | 47.71 |
| T2a | 66 064 221 | 65 936 595 | 61 819 035 | 97.94 | 94.03 | 47.93 |
| T2b | 65 163 741 | 65 017 603 | 61 516 931 | 97.48 | 93.01 | 47.94 |
| T2c | 62 902 745 | 62 794 761 | 58 409 372 | 98.06 | 94.28 | 48.21 |
| 样本 Sample | 0 ~ 0.1(%) | 0.1 ~ 1(%) | 1 ~ 10(%) | 10 ~ 100(%) | 100 ~ 1 000(%) | > 1 000(%) |
|---|---|---|---|---|---|---|
| T1a | 12 189(30.59) | 9 640(24.21) | 10 392(26.08) | 6 725(16.88) | 847(2.13) | 48(0.12) |
| T1b | 12 998(32.62) | 9 860(24.77) | 9 968(25.02) | 6 144(15.42) | 806(2.02) | 61(0.15) |
| T1c | 13 637(34.22) | 9 464(23.75) | 9 652(24.22) | 6 221(15.61) | 811(2.04) | 61(0.15) |
| T2a | 13 724(34.44) | 8 871(22.26) | 9 473(23.77) | 6 755(16.95) | 964(2.42) | 59(0.15) |
| T2b | 15 133(37.98) | 9 288(23.31) | 8 889(22.31) | 5 534(13.89) | 937(2.35) | 65(0.16) |
| T2c | 14 104(35.40) | 9 408(23.61) | 9 433(23.67) | 5 816(14.60) | 1 023(2.57) | 62(0.16) |
| 平均Mean | 13 631(34.21) | 9 422(23.65) | 9 635(24.18) | 6 199(15.56) | 898(2.26) | 59(0.15) |
表3 样本表达量FPKM数值及比例统计
Table 3 Sample expression volume FPKM value and proportion statistics
| 样本 Sample | 0 ~ 0.1(%) | 0.1 ~ 1(%) | 1 ~ 10(%) | 10 ~ 100(%) | 100 ~ 1 000(%) | > 1 000(%) |
|---|---|---|---|---|---|---|
| T1a | 12 189(30.59) | 9 640(24.21) | 10 392(26.08) | 6 725(16.88) | 847(2.13) | 48(0.12) |
| T1b | 12 998(32.62) | 9 860(24.77) | 9 968(25.02) | 6 144(15.42) | 806(2.02) | 61(0.15) |
| T1c | 13 637(34.22) | 9 464(23.75) | 9 652(24.22) | 6 221(15.61) | 811(2.04) | 61(0.15) |
| T2a | 13 724(34.44) | 8 871(22.26) | 9 473(23.77) | 6 755(16.95) | 964(2.42) | 59(0.15) |
| T2b | 15 133(37.98) | 9 288(23.31) | 8 889(22.31) | 5 534(13.89) | 937(2.35) | 65(0.16) |
| T2c | 14 104(35.40) | 9 408(23.61) | 9 433(23.67) | 5 816(14.60) | 1 023(2.57) | 62(0.16) |
| 平均Mean | 13 631(34.21) | 9 422(23.65) | 9 635(24.18) | 6 199(15.56) | 898(2.26) | 59(0.15) |
| 基因ID Gene ID | 花蕾期 Flower budding stage | 初开期Early flowering stage | 盛开期Blooming stage | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T1a- FPKM | T2a- FPKM | log2 (FC) | FDR | T1b- FPKM | T2b- FPKM | log2 (FC) | FDR | T1c- FPKM | T2c- FPKM | log2 (FC) | FDR | |
| Rc_0097351 | 0.9 | 33.1 | 5.1 | 5.2E-17 | 1.6 | 61.5 | 5.3 | 2.2E-68 | 0.9 | 61.0 | 6.1 | 3.1E-08 |
| Rc_0096771 | 123.6 | 415.8 | 1.8 | 2.7E-11 | 141.0 | 453.8 | 1.7 | 3.6E-12 | 74.0 | 223.5 | 1.6 | 2.2E-03 |
| Rc_0199551 | 30.1 | 91.2 | 1.6 | 2.4E-02 | 21.0 | 200.4 | 3.3 | 8.6E-13 | 17.1 | 360.2 | 4.4 | 3.7E-15 |
| Rc_0131551 | 9.4 | 2.2 | -2.1 | 4.2E-06 | 12.1 | 0.8 | -3.9 | 3.1E-12 | 7.8 | 0.9 | -3.2 | 1.0E-04 |
| Rc_0014811 | 30.2 | 1.8 | -4.1 | 1.1E-31 | 24.7 | 1.2 | -4.4 | 3.5E-31 | 29.7 | 1.1 | -4.8 | 3.3E-17 |
| Rc_0110711 | 541.7 | 227.9 | -1.2 | 8.0E-03 | 753.1 | 235.4 | -1.7 | 7.1E-04 | 676.7 | 152.4 | -2.2 | 6.1E-03 |
| Rc_0026721 | 0.1 | 32.0 | 8.5 | 9.7E-22 | 0.3 | 41.7 | 7.3 | 1.8E-42 | 0.3 | 29.5 | 6.8 | 4.8E-06 |
| Rc_0017561 | 0.4 | 15.3 | 5.3 | 7.3E-17 | 0.4 | 29.1 | 6.3 | 4.0E-39 | 0 | 18.0 | 8.9 | 6.4E-11 |
| Rc_0469861 | 23.8 | 55.5 | 1.2 | 6.0E-03 | 10.4 | 55.4 | 2.4 | 5.7E-04 | 6.7 | 43.4 | 2.7 | 1.3E-02 |
| Rc_0068591 | 23.5 | 48.3 | 1.0 | 3.4E-02 | 12.8 | 60.6 | 2.2 | 5.7E-17 | 6.1 | 31.1 | 2.4 | 8.0E-03 |
| Rc_0496721 | 38.6 | 6.2 | -2.6 | 3.4E-06 | 76.1 | 4.8 | -4.0 | 2.4E-09 | 26.2 | 4.4 | -2.6 | 3.5E-04 |
| Rc_0077971 | 71.6 | 0.1 | -10.0 | 6.7E-05 | 38.8 | 0.2 | -7.8 | 4.7E-09 | 5.3 | 0.6 | -3.3 | 2.1E-02 |
| Rc_0188031 | 13.7 | 5.7 | -1.3 | 4.6E-04 | 4.8 | 1.8 | -1.4 | 2.3E-02 | 6.4 | 0.8 | -3.0 | 4.1E-03 |
| Rc_0017641 | 12.6 | 0.7 | -4.2 | 1.9E-04 | 7.8 | 0.3 | -4.6 | 8.7E-05 | 23.9 | 0.4 | -6.1 | 1.0E-07 |
| Rc_0430481 | 7.6 | 0.1 | -6.6 | 4.1E-07 | 14.4 | 0.3 | -5.7 | 5.0E-14 | 4.4 | 0 | -12.1 | 1.7E-03 |
| Rc_0179641 | 372.9 | 0.4 | -9.8 | 1.5E-06 | 197.1 | 0.3 | -9.4 | 6.3E-12 | 26.6 | 1.6 | -4.0 | 2.8E-02 |
| Rc_0433321 | 565.7 | 0.3 | -10.7 | 1.7E-03 | 318.9 | 0.3 | -10.3 | 1.4E-07 | 41.8 | 2.3 | -4.2 | 1.2E-02 |
| Rc_0101451 | 62.8 | 22.8 | -1.5 | 5.2E-03 | 66.9 | 8.5 | -3.0 | 1.3E-04 | 33.3 | 2.9 | -3.5 | 3.8E-09 |
| Rc_0101491 | 154.1 | 57.6 | -1.4 | 9.1E-03 | 156.5 | 25.5 | -2.6 | 2.8E-04 | 95.2 | 10.6 | -3.2 | 1.4E-10 |
| Rc_0186071 | 33.1 | 68.3 | 1.0 | 2.8E-03 | 11.0 | 51.0 | 2.2 | 1.6E-09 | 4.9 | 23.7 | 2.3 | 1.3E-03 |
| Rc_0071571 | 197.3 | 26.0 | -2.9 | 1.0E-04 | 269.3 | 20.2 | -3.7 | 5.0E-08 | 100.8 | 14.7 | -2.8 | 8.0E-04 |
| Rc_0226301 | 95.8 | 9.8 | -3.3 | 3.4E-16 | 37.7 | 4.3 | -3.1 | 9.2E-09 | 28.6 | 1.8 | -4.0 | 4.8E-10 |
| Rc_0353511 | 2.6 | 45.2 | 4.1 | 4.6E-11 | 2.6 | 24.8 | 3.3 | 1.8E-08 | 0.9 | 6.1 | 2.7 | 1.1E-02 |
| Rc_0137231 | 0.2 | 53.2 | 8.0 | 2.2E-24 | 0.3 | 27.7 | 6.8 | 1.3E-36 | 0.1 | 12.2 | 7.7 | 4.8E-10 |
| Rc_0283821 | 43.4 | 315.4 | 2.9 | 5.1E-03 | 7.6 | 36.7 | 2.3 | 2.9E-08 | 5.4 | 36.9 | 2.8 | 5.0E-02 |
| Rc_0059511 | 91.7 | 2 062.3 | 4.5 | 1.1E-03 | 65.3 | 2 952.4 | 5.5 | 2.2E-10 | 38.8 | 2 143.2 | 5.8 | 1.3E-12 |
| Rc_0212441 | 95.4 | 2.9 | -5.0 | 3.2E-08 | 470.5 | 44.3 | -3.4 | 8.9E-09 | 993.0 | 98.3 | -3.3 | 8.7E-04 |
表4 3个开花阶段大花香水月季(T1)和单瓣香水月季(T2)花香通路差异表达基因表达情况
Table 4 Expression of differentially expressed genes in floral pathway of Rosa odorata var. gigantea(T1)and R. odorata var. odorata(T2)at the same flowering stage
| 基因ID Gene ID | 花蕾期 Flower budding stage | 初开期Early flowering stage | 盛开期Blooming stage | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T1a- FPKM | T2a- FPKM | log2 (FC) | FDR | T1b- FPKM | T2b- FPKM | log2 (FC) | FDR | T1c- FPKM | T2c- FPKM | log2 (FC) | FDR | |
| Rc_0097351 | 0.9 | 33.1 | 5.1 | 5.2E-17 | 1.6 | 61.5 | 5.3 | 2.2E-68 | 0.9 | 61.0 | 6.1 | 3.1E-08 |
| Rc_0096771 | 123.6 | 415.8 | 1.8 | 2.7E-11 | 141.0 | 453.8 | 1.7 | 3.6E-12 | 74.0 | 223.5 | 1.6 | 2.2E-03 |
| Rc_0199551 | 30.1 | 91.2 | 1.6 | 2.4E-02 | 21.0 | 200.4 | 3.3 | 8.6E-13 | 17.1 | 360.2 | 4.4 | 3.7E-15 |
| Rc_0131551 | 9.4 | 2.2 | -2.1 | 4.2E-06 | 12.1 | 0.8 | -3.9 | 3.1E-12 | 7.8 | 0.9 | -3.2 | 1.0E-04 |
| Rc_0014811 | 30.2 | 1.8 | -4.1 | 1.1E-31 | 24.7 | 1.2 | -4.4 | 3.5E-31 | 29.7 | 1.1 | -4.8 | 3.3E-17 |
| Rc_0110711 | 541.7 | 227.9 | -1.2 | 8.0E-03 | 753.1 | 235.4 | -1.7 | 7.1E-04 | 676.7 | 152.4 | -2.2 | 6.1E-03 |
| Rc_0026721 | 0.1 | 32.0 | 8.5 | 9.7E-22 | 0.3 | 41.7 | 7.3 | 1.8E-42 | 0.3 | 29.5 | 6.8 | 4.8E-06 |
| Rc_0017561 | 0.4 | 15.3 | 5.3 | 7.3E-17 | 0.4 | 29.1 | 6.3 | 4.0E-39 | 0 | 18.0 | 8.9 | 6.4E-11 |
| Rc_0469861 | 23.8 | 55.5 | 1.2 | 6.0E-03 | 10.4 | 55.4 | 2.4 | 5.7E-04 | 6.7 | 43.4 | 2.7 | 1.3E-02 |
| Rc_0068591 | 23.5 | 48.3 | 1.0 | 3.4E-02 | 12.8 | 60.6 | 2.2 | 5.7E-17 | 6.1 | 31.1 | 2.4 | 8.0E-03 |
| Rc_0496721 | 38.6 | 6.2 | -2.6 | 3.4E-06 | 76.1 | 4.8 | -4.0 | 2.4E-09 | 26.2 | 4.4 | -2.6 | 3.5E-04 |
| Rc_0077971 | 71.6 | 0.1 | -10.0 | 6.7E-05 | 38.8 | 0.2 | -7.8 | 4.7E-09 | 5.3 | 0.6 | -3.3 | 2.1E-02 |
| Rc_0188031 | 13.7 | 5.7 | -1.3 | 4.6E-04 | 4.8 | 1.8 | -1.4 | 2.3E-02 | 6.4 | 0.8 | -3.0 | 4.1E-03 |
| Rc_0017641 | 12.6 | 0.7 | -4.2 | 1.9E-04 | 7.8 | 0.3 | -4.6 | 8.7E-05 | 23.9 | 0.4 | -6.1 | 1.0E-07 |
| Rc_0430481 | 7.6 | 0.1 | -6.6 | 4.1E-07 | 14.4 | 0.3 | -5.7 | 5.0E-14 | 4.4 | 0 | -12.1 | 1.7E-03 |
| Rc_0179641 | 372.9 | 0.4 | -9.8 | 1.5E-06 | 197.1 | 0.3 | -9.4 | 6.3E-12 | 26.6 | 1.6 | -4.0 | 2.8E-02 |
| Rc_0433321 | 565.7 | 0.3 | -10.7 | 1.7E-03 | 318.9 | 0.3 | -10.3 | 1.4E-07 | 41.8 | 2.3 | -4.2 | 1.2E-02 |
| Rc_0101451 | 62.8 | 22.8 | -1.5 | 5.2E-03 | 66.9 | 8.5 | -3.0 | 1.3E-04 | 33.3 | 2.9 | -3.5 | 3.8E-09 |
| Rc_0101491 | 154.1 | 57.6 | -1.4 | 9.1E-03 | 156.5 | 25.5 | -2.6 | 2.8E-04 | 95.2 | 10.6 | -3.2 | 1.4E-10 |
| Rc_0186071 | 33.1 | 68.3 | 1.0 | 2.8E-03 | 11.0 | 51.0 | 2.2 | 1.6E-09 | 4.9 | 23.7 | 2.3 | 1.3E-03 |
| Rc_0071571 | 197.3 | 26.0 | -2.9 | 1.0E-04 | 269.3 | 20.2 | -3.7 | 5.0E-08 | 100.8 | 14.7 | -2.8 | 8.0E-04 |
| Rc_0226301 | 95.8 | 9.8 | -3.3 | 3.4E-16 | 37.7 | 4.3 | -3.1 | 9.2E-09 | 28.6 | 1.8 | -4.0 | 4.8E-10 |
| Rc_0353511 | 2.6 | 45.2 | 4.1 | 4.6E-11 | 2.6 | 24.8 | 3.3 | 1.8E-08 | 0.9 | 6.1 | 2.7 | 1.1E-02 |
| Rc_0137231 | 0.2 | 53.2 | 8.0 | 2.2E-24 | 0.3 | 27.7 | 6.8 | 1.3E-36 | 0.1 | 12.2 | 7.7 | 4.8E-10 |
| Rc_0283821 | 43.4 | 315.4 | 2.9 | 5.1E-03 | 7.6 | 36.7 | 2.3 | 2.9E-08 | 5.4 | 36.9 | 2.8 | 5.0E-02 |
| Rc_0059511 | 91.7 | 2 062.3 | 4.5 | 1.1E-03 | 65.3 | 2 952.4 | 5.5 | 2.2E-10 | 38.8 | 2 143.2 | 5.8 | 1.3E-12 |
| Rc_0212441 | 95.4 | 2.9 | -5.0 | 3.2E-08 | 470.5 | 44.3 | -3.4 | 8.9E-09 | 993.0 | 98.3 | -3.3 | 8.7E-04 |
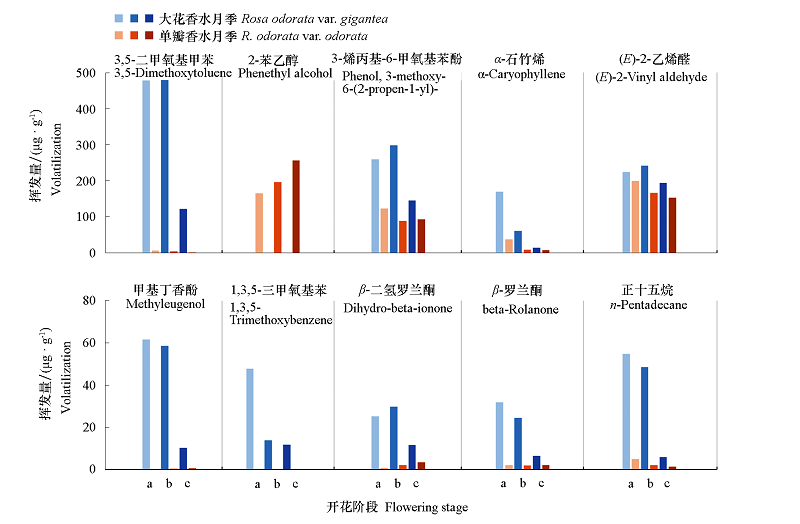
图8 主要差异花香物质释放量 a. 花蕾期;b. 初开期;c. 全开期。
Fig. 8 Main difference in the release of floral substance a:Flower budding stage;b:Early flowering stage;c:Blooming stage.
| [1] | Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp:an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics, 34 (17):i884-i890. |
| [2] |
Davis A P, Sherlock G, Botstein D, Eppig J T, Matese J C, Harris M A, Kasarskis A, Blake J A, Dolinski K, Dwight S S, Issel-Tarver L, Ringwald M, Ball C A, Richardson J E, Rubin G M, Cherry J M, Ashburner M, Lewis S, Butler H, Hill D P. 2000. Gene Ontology:tool for the unification of biology. Nature Genetics, 25 (1):25-29.
pmid: 10802651 |
| [3] |
Dexter R, Qualley A, Kish C M, Ma C J, Koeduka T, Nagegowda D A, Dudareva N, Pichersky E, Clark D. 2007. Characterization of a petunia acetyltransferase involved in the biosynthesis of the floral volatile isoeugenol. Plant Journal, 49 (2):265-275.
doi: 10.1111/j.1365-313X.2006.02954.x URL |
| [4] | Fan Li. 2013. Bioinformatic and functional analysis of mulberry genes involved in lignin biosynthesis[Ph. D. Dissertation]. Chongqing:Southwest University. (in Chinese) |
| 范丽. 2013. 桑树木质素合成基因的生物信息和功能分析[博士论文]. 重庆: 西南大学. | |
| [5] | Guo Guang-yan, Bo Feng, Liu Wei, Mi Cai-li. 2015. Advances in research of the regulation of transcription factors of lignin biosynthesis. Scientia Agricultura Sinica, 48 (7):1277-1287. (in Chinese) |
| 郭光艳, 柏峰, 刘伟, 秘彩莉. 2015. 转录因子对木质素生物合成调控的研究进展. 中国农业科学, 48 (7):1277-1287. | |
| [6] |
Kanehisa M. 2000. KEGG:Kyoto encyclopedia of genes and genomes. Nucleic Acids Research, 28 (1):27-30.
pmid: 10592173 |
| [7] |
Kim D, Langmead B, Salzberg S L. 2015. HISAT:a fast spliced aligner with low memory requirements. Nature Methods, 12 (4):357-360.
doi: 10.1038/NMETH.3317 |
| [8] |
Langmead B, Salzberg S L. 2012. Fast gapped-read alignment with Bowtie 2. Nature Methods, 9 (4):357-359.
doi: 10.1038/nmeth.1923 pmid: 22388286 |
| [9] |
Lavid N, Wang J, Shalit M, Guterman I, Bar E, Beuerle T, Menda N, Shafir S, Zamir D, Adam Z, Vainstein A, Weiss D, Pichersky E, Lewinsohn E. 2002. O-Methyltransferases involved in the biosynthesis of volatile phenolic derivatives in rose petals. Plant Physiology (Bethesda), 129 (4):1899-1907.
doi: 10.1104/pp.005330 URL |
| [10] | Li Jin-hua, Yan Hui-jun, Yang Jin-hong, Jian Hong-ying, Zhang Hao, Chen Min, Tang Kai-xue. 2018. Analysis of volatile components from Rosa odorata complex by SPME-GC/MS. Southwest China Journal of Agricultural Sciences, 31 (3):587-591. (in Chinese) |
| 李晋华, 晏慧君, 杨锦红, 蹇洪英, 张颢, 陈敏, 唐开学. 2018. 香水月季复合群(Rosa odorata Complex)花香成分分析. 西南农业学报, 31 (3):587-591. | |
| [11] | Liu Wei. 2019. Functional research of CmCADs in lignin biosynthesis in oriental melon(Cucumis melo L.)under abiotic stresses[Ph. D. Dissertation]. Shenyang:Shenyang Agricultural University. (in Chinese) |
| 刘威. 2019. 薄皮甜瓜肉桂醇脱氢酶(CAD)在非生物胁迫下参与木质素合成的功能探究[博士论文]. 沈阳: 沈阳农业大学. | |
| [12] |
Love M I, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology, 15 (12):550.
doi: 10.1186/s13059-014-0550-8 URL |
| [13] | Ma Zhuan-zhuan, Pang Xiao-qing, Chen Rong, Pei Xiao-lin, Wang Qiu-yan, Xie Tian, Yin Xiao-pu. 2015. Research progress of key enzymes in biosynthesis pathway of terpenes. Journal of Hangzhou Normal University(Natural Science Edition), 14 (6):608-615. (in Chinese) |
| 马转转, 庞潇卿, 谌容, 裴晓林, 王秋岩, 谢恬, 殷晓浦. 2015. 萜类化合物生物合成途径中关键酶的研究进展. 杭州师范大学学报(自然科学版), 14 (6):608-615. | |
| [14] | Min Dan-dan, Tang Mei-qiong, Li Gang, Qu Xiao-sheng, Mou Jian-hua. 2015. Cloning and expression analysis of geranylgeranyl diphosphate synthase gene of Panax notoginseng. Chinese Journal of Traditional Chinese Medicine, 40 (11):2090-2095. (in Chinese) |
| 闵丹丹, 唐美琼, 李刚, 屈啸声, 缪剑华. 2015. 三七香叶基香叶基焦磷酸合酶基因的克隆及表达分析. 中国中药杂志, 40 (11):2090-2095. | |
| [15] |
Mortazavi A, Williams B A, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods, 5 (7):621-628.
doi: 10.1038/nmeth.1226 pmid: 18516045 |
| [16] |
Pertea M, Pertea G M, Antonescu C M, Chang T, Mendell J T, Salzberg S L. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nature Biotechnology, 33 (3):290-295.
doi: 10.1038/nbt.3122 URL |
| [17] |
Raymond O, Gouzy J, Just J, Badouin H, Verdenaud M, Lemainque A, Vergne P, Moja S, Choisne N, Pont C, Carrere S, Caissard J, Couloux A, Cottret L, Aury J, Szecsi J, Latrasse D, Madoui M, Francois L, Fu X, Yang S, Dubois A, Piola F, Larrieu A, Perez M, Labadie K, Perrier L, Govetto B, Labrousse Y, Villand P, Bardoux C, Boltz V, Lopez-Roques C, Heitzler P, Vernoux T, Vandenbussche M, Quesneville H, Boualem A, Bendahmane A, Liu C, Le Bris M, Salse J, Baudino S, Benhamed M, Wincker P, Bendahmane M. 2018. The Rosa genome provides new insights into the domestication of modern roses. Nature Genetics, 50 (6):772.
doi: 10.1038/s41588-018-0110-3 URL |
| [18] |
Scalliet G, Journot N, Jullien F, Baudino S, Magnard J L, Channeliere S, Vergne P, Dumas C, Bendahmane M, Cock J M, Hugueney P. 2002. Biosynthesis of the major scent components 3,5-dimethoxytoluene and 1,3,5-trimethoxybenzene by novel rose O-methyltransferases. FEBS Letters, 523 (1-3):113-118.
pmid: 12123815 |
| [19] | Tang Yun. 2019. Molecular identification of the biosynthesis pathway of sesquiterpenes and functional analysis of PatFPPS[Ph. D. Disseration]. Guangzhou:Guangzhou University of Chinese Medica. (in Chinese) |
| 唐云. 2019. 广藿香倍半萜合成通路关键酶基因鉴定和PatFPPS的功能分析[博士论文]. 广州: 广州中医药大学. | |
| [20] | Wang Hua-mei, Yu Yan-chong, Fu Chun-xiang, Zhou Gong-ke, Gao Huan-huan. 2014. Research progress of caffeoyl-CoA 3-O-methyltransferase key enzyme in Lignin synthesis. Genomics and Applied Biology, 33 (2):458-466. (in Chinese) |
| 王华美, 于延冲, 付春祥, 周功克, 高欢欢. 2014. 木质素合成关键酶咖啡酰辅酶A氧甲基转移酶的研究进展. 基因组学与应用生物学, 33 (2):458-466. | |
| [21] | Wang Jia. 2016. Construction of over-expression vector of RrDXR and RrAAT and genetic transformaton study in Rosa rugosa[Ph. D. Dissertation]. Yangzhou:Yangzhou University. (in Chinese) |
| 王佳. 2016. 玫瑰花香相关基因RrDXR和RrAAT超表达载体构建及遗传转化研究[博士论文]. 扬州: 扬州大学. | |
| [22] | Wang Jian-yong. 2014. The study on cloning and function of seven genes involved in the fatty acid metabolism of Camellia oleifera seeds[Ph. D. Dissertation]. Changsha: Central South University of Forestry and Technology. (in Chinese) |
| 王建勇. 2014. 油茶种子脂肪酸代谢过程7个关键酶基因的克隆与功能研究[博士论文]. 长沙: 中南林业科技大学. | |
| [23] | Wu Shao-yan, Li Jin, Zhou Shun, Lu Xiao-ying. 2019. Study on synthesis and application of coumarin and its derivatives. Guangdong Chemical Industry, 46 (24):127-128. (in Chinese) |
| 吴绍艳, 李进, 周顺, 卢小英. 2019. 香豆素及其衍生物的合成及应用研究. 广东化工, 46 (24):127-128. | |
| [24] |
Wu S, Watanabe N, Mita S, Dohra H. 2004. The key role of phloroglucinol O-Methyltransferase in the biosynthesis of Rosa chinensis volatile 1,3,5-trimethoxybenzene1. Plant Physiology (Bethesda), 135 (1):95-102.
doi: 10.1104/pp.103.037051 URL |
| [25] | Zhang Xu, Wang Xiao-jia, Li Si-chen, Dong Tian-tian, Wang Zhi-hui. 2019. Research progress of lignin biosynthesis and regulation during granulation of citrus. Acta Agriculturae Zhejiangensis, 31 (12):2131-2140. (in Chinese) |
| 张旭, 王小佳, 黎思辰, 董甜甜, 汪志辉. 2019. 柑橘果实粒化过程中木质素生物合成与调控研究进展. 浙江农业学报, 31 (12):2131-2140. | |
| [26] | Zhou Han-chen, Lei Pan-deng, Ding Yong. 2016. Research advance on β-glucosidase of tea plant. Journal of Tea Science, 36 (2):111-118. (in Chinese) |
| 周汉琛, 雷攀登, 丁勇. 2016. 茶树β-葡萄糖苷酶研究进展. 茶叶科学, 36 (02):111-118. | |
| [27] |
Zhou L, Yu C, Cheng B, Han Y, Luo L, Pan H, Zhang Q. 2020a. Studies on the volatile compounds in flower extracts of Rosa odorata and R. chinensis. Industrial Crops and Products, 146:112143.
doi: 10.1016/j.indcrop.2020.112143 URL |
| [28] |
Zhou L, Yu C, Cheng B, Wan H, Luo L, Pan H, Zhang Q. 2020b. Volatile compound analysis and aroma evaluation of tea-scented roses in China. Industrial Crops and Products, 155:112735.
doi: 10.1016/j.indcrop.2020.112735 URL |
| [1] | 欧阳丽莹, 黄艳飞, 陈君梅, 阳 淑, 练华山. 微型月季新品种‘九儿’[J]. 园艺学报, 2022, 49(S2): 223-224. |
| [2] | 练华山, 欧阳丽莹, 吴 珊, 陈君梅, 舒 彬. 微型月季新品种‘夜华’[J]. 园艺学报, 2022, 49(S2): 225-226. |
| [3] | 陈君梅, 练华山, 黄艳飞, 禹 婷, 蒋天仪. 微型月季新品种‘流星’[J]. 园艺学报, 2022, 49(S2): 227-228. |
| [4] | 贾鑫, 曾臻, 陈月, 冯慧, 吕英民, 赵世伟. 月季‘月月粉’RcDREB2A的克隆与表达分析[J]. 园艺学报, 2022, 49(9): 1945-1956. |
| [5] | 李茂福, 杨媛, 王华, 范又维, 孙佩, 金万梅. 月季中R2R3-MYB基因RhMYB113c调控花青素苷合成[J]. 园艺学报, 2022, 49(9): 1957-1966. |
| [6] | 李茂福, 杨媛, 王华, 范又维, 孙佩, 金万梅. 月季自交不亲和性S-RNase的鉴定与分析[J]. 园艺学报, 2022, 49(1): 157-165. |
| [7] | 周 群. 三角梅新品种‘香妃1号’[J]. 园艺学报, 2021, 48(S2): 2991-2992. |
| [8] | 李贵生. 猕猴桃‘金艳'和‘红阳'果实转录组的比较分析[J]. 园艺学报, 2021, 48(6): 1183-1196. |
| [9] | 温佳辛, 王超林, 冯慧, 李珊珊, 王亮生, 武荣花, 赵世伟. 月季花色研究进展[J]. 园艺学报, 2021, 48(10): 2044-2056. |
| [10] | 吴洪娥, 金 晶, 吴 楠, 董万鹏, 朱 立, 周洪英. 丰花月季新品种‘花好月圆’[J]. 园艺学报, 2020, 47(S2): 3077-3078. |
| [11] | 丁爱琴1,2,韩坤桀1,杨立晨1,张竹君1,耿子雯1,付鲁峰1,刘庆华1,*,姜新强1,*. 月季RhNAC31互作蛋白的筛选及分析[J]. 园艺学报, 2020, 47(7): 1335-1344. |
| [12] | 王 焕*,郑日如*,曹声海,张 通,史若明,王彩云,罗 靖**. 月季花瓣特异表达启动子的筛选和鉴定[J]. 园艺学报, 2020, 47(4): 686-698. |
| [13] | 高述民 1,周 燕 1,2,*,范莉娟 1,2,巢 阳 2. 微型月季新品种‘桔月’[J]. 园艺学报, 2020, 47(2): 401-402. |
| [14] | 王丽花1,段金辉2,*,李清云2,段云晟2,薛祖旺2,范荣旭2. 月季新品种‘金焰’[J]. 园艺学报, 2020, 47(1): 199-200. |
| [15] | 李升娟,许忠民*,郭 佳,张恩慧,姜 娇,石汶汶. 基于叶球转录组数据比较的甘蓝杂种优势分析[J]. 园艺学报, 2019, 46(6): 1079-1092. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司