
园艺学报 ›› 2021, Vol. 48 ›› Issue (11): 2251-2261.doi: 10.16420/j.issn.0513-353x.2021-0397
杨婷1, 薛珍珍1, 李娜2, 郎校安2, 李凌飞2,*( ), 钟春梅1,*(
), 钟春梅1,*( )
)
收稿日期:2021-05-17
修回日期:2021-09-15
发布日期:2021-12-02
通讯作者:
李凌飞,钟春梅
E-mail:lingfei_li@szbg.ac.cn;zhongchunmei@scau.edu.cn
基金资助:
YANG Ting1, XUE Zhenzhen1, LI Na2, LANG Xiaoan2, LI Lingfei2,*( ), ZHONG Chunmei1,*(
), ZHONG Chunmei1,*( )
)
Received:2021-05-17
Revised:2021-09-15
Published:2021-12-02
Contact:
LI Lingfei,ZHONG Chunmei
E-mail:lingfei_li@szbg.ac.cn;zhongchunmei@scau.edu.cn
摘要:
为研究铁十字秋海棠(Begonia masoniana)斑叶发育过程中最适内参基因与花青素苷合成相关基因的表达,以铁十字秋海棠7个时期的叶片为材料,通过实时荧光定量PCR技术、GeNorm、NormFinder和BestKeeper对11个候选内参基因(PP2A、EF1α、GADPH、CYP、elF4A、ACT2、18S、UBQ10、ACT7、PPR和HDH)进行表达稳定性评估,最适内参基因为ACT7和PP2A。分别以ACT7和PP2A为内参基因对6个花青素苷合成通路相关基因(CHS、F3H、F3’H、FLS、DFR和UFGT)的表达量进行分析,结果显示该通路相关基因表达先升后降,与叶片花青素苷含量变化趋势较为一致。
中图分类号:
杨婷, 薛珍珍, 李娜, 郎校安, 李凌飞, 钟春梅. 铁十字秋海棠斑叶发育过程内参基因筛选及验证[J]. 园艺学报, 2021, 48(11): 2251-2261.
YANG Ting, XUE Zhenzhen, LI Na, LANG Xiaoan, LI Lingfei, ZHONG Chunmei. Reference Genes Selection and Validation in Begonia masoniana Leaves of Different Developmental Stages[J]. Acta Horticulturae Sinica, 2021, 48(11): 2251-2261.
| 时期 Stage | 叶片长/cm Leaf length | 叶片宽/cm Leaf width | 生长天数/d Growth days | 特征描述 Feature description |
|---|---|---|---|---|
| S1 | 1.2 ~ 1.6 | 1.0 ~ 1.2 | 2 ~ 3 | 叶片卷曲,表皮毛较长,全叶呈红色 Leaf curly,hirsute-villous,whole leaf is red |
| S2 | 2.2 ~ 2.8 | 1.8 ~ 2.2 | 6 ~ 8 | 叶片舒张,表皮毛长,叶片质地较厚,全叶呈红色 Leaf expansion,hirsute-villous,thick,whole leaf is red |
| S3 | 4.1 ~ 5.1 | 3.1 ~ 3.7 | 10 ~ 12 | 叶斑初显,沿主叶脉规则分布,全叶呈红色 Leaf variegation appearing,distributed along the main vein,whole leaf is red |
| S4 | 6.8 ~ 7.4 | 4.6 ~ 6.2 | 15 ~ 18 | 叶斑颜色继续加深,叶斑以外部分叶色偏黄 Leaf variegation deepen,other parts of leaf show yellowish |
| S5 | 10.2 ~ 12.8 | 7.7 ~ 8.7 | 23 ~ 25 | 叶斑明显,叶斑以外部分叶色逐渐转绿 Leaf variegation begin obvious,other parts of leaf turn green |
| S6 | 12.7 ~ 15.0 | 9.2 ~ 11.9 | 33 ~ 35 | 叶片继续生长,叶斑以外部分叶色全绿 Leaf continues to grow,and other parts except leaf variegation are all green |
| S7 | 20.1 ~ 21.5 | 15.2 ~ 16.1 | 48 ~ 52 | 叶片大小达到最大 Leaf size reaches maximum |
表1 铁十字秋海棠叶片发育时期划分
Table 1 Developmental stages of leaves in Begonia masoniana
| 时期 Stage | 叶片长/cm Leaf length | 叶片宽/cm Leaf width | 生长天数/d Growth days | 特征描述 Feature description |
|---|---|---|---|---|
| S1 | 1.2 ~ 1.6 | 1.0 ~ 1.2 | 2 ~ 3 | 叶片卷曲,表皮毛较长,全叶呈红色 Leaf curly,hirsute-villous,whole leaf is red |
| S2 | 2.2 ~ 2.8 | 1.8 ~ 2.2 | 6 ~ 8 | 叶片舒张,表皮毛长,叶片质地较厚,全叶呈红色 Leaf expansion,hirsute-villous,thick,whole leaf is red |
| S3 | 4.1 ~ 5.1 | 3.1 ~ 3.7 | 10 ~ 12 | 叶斑初显,沿主叶脉规则分布,全叶呈红色 Leaf variegation appearing,distributed along the main vein,whole leaf is red |
| S4 | 6.8 ~ 7.4 | 4.6 ~ 6.2 | 15 ~ 18 | 叶斑颜色继续加深,叶斑以外部分叶色偏黄 Leaf variegation deepen,other parts of leaf show yellowish |
| S5 | 10.2 ~ 12.8 | 7.7 ~ 8.7 | 23 ~ 25 | 叶斑明显,叶斑以外部分叶色逐渐转绿 Leaf variegation begin obvious,other parts of leaf turn green |
| S6 | 12.7 ~ 15.0 | 9.2 ~ 11.9 | 33 ~ 35 | 叶片继续生长,叶斑以外部分叶色全绿 Leaf continues to grow,and other parts except leaf variegation are all green |
| S7 | 20.1 ~ 21.5 | 15.2 ~ 16.1 | 48 ~ 52 | 叶片大小达到最大 Leaf size reaches maximum |
| 基因 Gene | 引物序列(5′-3′) Primer sequence |
|---|---|
| CHS | F:TGCACCACAAGCGGAGTCGA;R:AGCAAGATCCTTGGCGACCCT |
| F3H | F:TTCCAGAACCCAGCGCCAGA;R:TCAGCCTGGCAAGCTCAAGGT |
| F3’H | F:GCGACTTCATTCCGGCGCTT;R:ACCGCCGATCCGCTTGTGTT |
| FLS | F:TGAGCAGCCCGGAATCACCA;R:TGTCGCACACCACACGTTCCT |
| DFR | F:AGACATTGGCAGAGCAGGCG;R:AGTGATCAGGCTCGGTGGCA |
| UFGT | F:TGAATGCCGCCCCAGAAAGCT;R:CGGCGAACCACAAGAAAGCGT |
表2 花青素苷合成途径中部分关键基因qRT-PCR的引物序列
Table 2 Primer sequences for some key genes in anthocyanin synthesis pathway derived from qRT-PCR
| 基因 Gene | 引物序列(5′-3′) Primer sequence |
|---|---|
| CHS | F:TGCACCACAAGCGGAGTCGA;R:AGCAAGATCCTTGGCGACCCT |
| F3H | F:TTCCAGAACCCAGCGCCAGA;R:TCAGCCTGGCAAGCTCAAGGT |
| F3’H | F:GCGACTTCATTCCGGCGCTT;R:ACCGCCGATCCGCTTGTGTT |
| FLS | F:TGAGCAGCCCGGAATCACCA;R:TGTCGCACACCACACGTTCCT |
| DFR | F:AGACATTGGCAGAGCAGGCG;R:AGTGATCAGGCTCGGTGGCA |
| UFGT | F:TGAATGCCGCCCCAGAAAGCT;R:CGGCGAACCACAAGAAAGCGT |
| 基因 Gene | 基因名称 Gene name | 引物序列(5′-3′) Primer sequence | 斜率 k | 扩增效率/% E | 相关系数 R2 |
|---|---|---|---|---|---|
| PP2A | Serine/threonine-PP2A catalytic subunit | F:TCCTGATGGAGTGCAAGCCGT R:ACAGACTGTCACCGGGCACT | -3.31 | 100.50 | 0.9984 |
| EF1α | Elongation factor 1α | F:TGCAGCTTGGCTCGGCAGAT R:TGGCAGGGGCGTGATCTAACA | -3.41 | 96.45 | 0.9996 |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | F:TGACGTTGTGGCAGCTCAGGT R:TCCGGCCAATGCGTCCGAAT | -3.32 | 100.08 | 0.9961 |
| CYP | Cyclophilin | F:ACGGTGCCAAGTTTGCCGAC R:AACTGTGAGCCGTTGGTTCCG | -3.34 | 99.25 | 0.9988 |
| elF4A | Eukaryotic translation initiation factor 4A-1 | F:ACCTCCATCGAATCGGCCGT R:TGCTGGCAACTCCTCCACCA | -3.20 | 105.35 | 0.9995 |
| ACT2 | Actin 2 | F:TTGCTCACAGAGGCACCGCT R:ACCGGTAGTACGACCACTGGCA | -3.33 | 99.66 | 0.9935 |
| 18S | 18S Ribosomal RNA | F:TGACGGATCGCACGGCCTTT R:TTCTCCGTCACCCGTCACCA | -3.45 | 94.92 | 0.9966 |
| UBQ10 | Polyubiquitin 10 | F:ACGCTTCGAGGTAGGTTTCTTGT R:ACGCATCGAAAACAACAACCGCA | -3.50 | 93.07 | 0.9991 |
| ACT7 | Actin 7 | F:TCGTGCTTGGAGGTTCGGCT R:CCAGCCTTCACCATTCCAGTTCCA | -3.27 | 102.21 | 0.9964 |
| PPR | Pentatricopeptide repeat superfamily protein | F:AGAAGTGCGAGTGGCTACCT R:CACAAACCCGCCGATCAGAGT | -3.19 | 105.82 | 0.9996 |
| HDH | Histidinol dehydrogenase | F:ATGTCCCTGGGGGTACTGCTGT R:TGCCATCCTGACTTGGGGGA | -3.36 | 98.44 | 0.9997 |
表3 铁十字秋海棠11个候选内参基因qRT-PCR的引物序列和扩增参数
Table 3 Primer sequences and amplification parameters for 11 candidate reference genes of Begonia masoniana
| 基因 Gene | 基因名称 Gene name | 引物序列(5′-3′) Primer sequence | 斜率 k | 扩增效率/% E | 相关系数 R2 |
|---|---|---|---|---|---|
| PP2A | Serine/threonine-PP2A catalytic subunit | F:TCCTGATGGAGTGCAAGCCGT R:ACAGACTGTCACCGGGCACT | -3.31 | 100.50 | 0.9984 |
| EF1α | Elongation factor 1α | F:TGCAGCTTGGCTCGGCAGAT R:TGGCAGGGGCGTGATCTAACA | -3.41 | 96.45 | 0.9996 |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | F:TGACGTTGTGGCAGCTCAGGT R:TCCGGCCAATGCGTCCGAAT | -3.32 | 100.08 | 0.9961 |
| CYP | Cyclophilin | F:ACGGTGCCAAGTTTGCCGAC R:AACTGTGAGCCGTTGGTTCCG | -3.34 | 99.25 | 0.9988 |
| elF4A | Eukaryotic translation initiation factor 4A-1 | F:ACCTCCATCGAATCGGCCGT R:TGCTGGCAACTCCTCCACCA | -3.20 | 105.35 | 0.9995 |
| ACT2 | Actin 2 | F:TTGCTCACAGAGGCACCGCT R:ACCGGTAGTACGACCACTGGCA | -3.33 | 99.66 | 0.9935 |
| 18S | 18S Ribosomal RNA | F:TGACGGATCGCACGGCCTTT R:TTCTCCGTCACCCGTCACCA | -3.45 | 94.92 | 0.9966 |
| UBQ10 | Polyubiquitin 10 | F:ACGCTTCGAGGTAGGTTTCTTGT R:ACGCATCGAAAACAACAACCGCA | -3.50 | 93.07 | 0.9991 |
| ACT7 | Actin 7 | F:TCGTGCTTGGAGGTTCGGCT R:CCAGCCTTCACCATTCCAGTTCCA | -3.27 | 102.21 | 0.9964 |
| PPR | Pentatricopeptide repeat superfamily protein | F:AGAAGTGCGAGTGGCTACCT R:CACAAACCCGCCGATCAGAGT | -3.19 | 105.82 | 0.9996 |
| HDH | Histidinol dehydrogenase | F:ATGTCCCTGGGGGTACTGCTGT R:TGCCATCCTGACTTGGGGGA | -3.36 | 98.44 | 0.9997 |
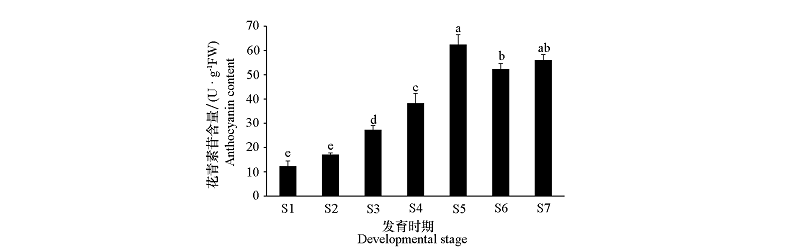
图2 铁十字秋海棠叶片不同时期的花青素苷含量 不同小写字母表示不同时期间差异显著(P < 0.05)。
Fig. 2 Anthocyanin content from Begonia masoniana leaves of different developmental stages Different lowercase letters indicate significant differences among different periods at 0.05 level.
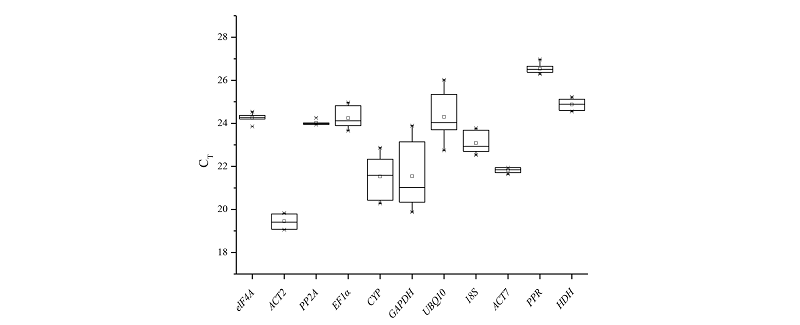
图5 11个内参候选基因的CT值分布图 中间横线是中位数,□是平均数,*是异常值。
Fig. 5 CT value distribution of 11 candidate reference genes The middle horizontal line is the median,□ is the average,* is the abnormal value.
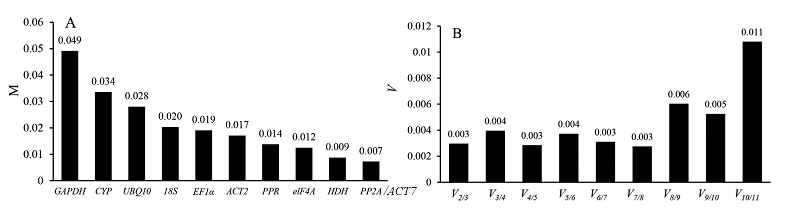
图6 GeNorm分析11个候选内参基因表达稳定性(A)和最佳内参基因数量(B)
Fig. 6 Expression stabilities of 11 candidate reference genes(A)andsoptimal numbers of reference genes(B)analyzed by GeNorm
| 基因 Gene | 几何平均数 GM | 算数平均数 AM | 最小值 Min | 最大值 Max | 标准偏差 SD | 稳定性排序 Stability rank |
|---|---|---|---|---|---|---|
| PP2A | 24.02 | 24.02 | 23.94 | 24.26 | 0.07 | 1 |
| ACT7 | 21.82 | 21.82 | 21.64 | 21.94 | 0.10 | 2 |
| elF4A | 24.26 | 24.26 | 23.86 | 24.54 | 0.14 | 3 |
| PPR | 26.54 | 26.54 | 26.29 | 26.97 | 0.15 | 4 |
| HDH | 24.88 | 24.88 | 24.55 | 25.23 | 0.19 | 5 |
| ACT2 | 19.45 | 19.45 | 19.06 | 19.83 | 0.26 | 6 |
| EF1α | 24.24 | 24.25 | 23.66 | 24.97 | 0.37 | 7 |
| 18S | 23.08 | 23.09 | 22.53 | 23.77 | 0.42 | 8 |
| CYP | 21.51 | 21.53 | 20.28 | 22.87 | 0.77 | 9 |
| UBQ10 | 24.28 | 24.30 | 22.75 | 26.02 | 0.82 | 10 |
| GAPDH | 21.50 | 21.55 | 19.88 | 23.89 | 1.27 | 11 |
表4 BestKeeper分析11个候选内参基因表达稳定性
Table 4 Expression stabilities of eleven candidate reference genes analyzed by BestKeeper
| 基因 Gene | 几何平均数 GM | 算数平均数 AM | 最小值 Min | 最大值 Max | 标准偏差 SD | 稳定性排序 Stability rank |
|---|---|---|---|---|---|---|
| PP2A | 24.02 | 24.02 | 23.94 | 24.26 | 0.07 | 1 |
| ACT7 | 21.82 | 21.82 | 21.64 | 21.94 | 0.10 | 2 |
| elF4A | 24.26 | 24.26 | 23.86 | 24.54 | 0.14 | 3 |
| PPR | 26.54 | 26.54 | 26.29 | 26.97 | 0.15 | 4 |
| HDH | 24.88 | 24.88 | 24.55 | 25.23 | 0.19 | 5 |
| ACT2 | 19.45 | 19.45 | 19.06 | 19.83 | 0.26 | 6 |
| EF1α | 24.24 | 24.25 | 23.66 | 24.97 | 0.37 | 7 |
| 18S | 23.08 | 23.09 | 22.53 | 23.77 | 0.42 | 8 |
| CYP | 21.51 | 21.53 | 20.28 | 22.87 | 0.77 | 9 |
| UBQ10 | 24.28 | 24.30 | 22.75 | 26.02 | 0.82 | 10 |
| GAPDH | 21.50 | 21.55 | 19.88 | 23.89 | 1.27 | 11 |
| [1] |
Andersen C L, Jensen J L, Orntoft T F. 2004. Normalization of real-time quantitative reverse transcription-PCR data:a model-based variance estimation approach to identify genes suited for normalization,applied to bladder and colon cancer data sets. Cancer Res, 64 (15):5245-5250.
doi: 10.1158/0008-5472.CAN-04-0496 URL |
| [2] | Bai Xue. 2020. Study on genes and substances related to anthocyanin biosynthesis by low temperature and high light stress in the leaves of Begonia semperflorens[M. D. Dissertation]. Zhengzhou: Henan Agricultural University. (in Chinese) |
| 白雪. 2020. 四季秋海棠叶片在低温和高光胁迫下次生花色素苷生物合成相关基因以及物质的研究[硕士论文]. 郑州: 河南农业大学. | |
| [3] |
Bustin S A. 2002. Quantification of mRNA using real-time reverse transcription PCR(RT-PCR):trends and problems. J Mol Endocrinol, 29 (1):23-39.
pmid: 12200227 |
| [4] | Cui Weihua, Guan Kaiyun. 2013. Diversity of leaf variegation in Chinese Begonias. Plant Diversity and Resources, 35 (2):119-127. (in Chinese) |
| 崔卫华, 管开云. 2013. 中国秋海棠属植物叶片斑纹多样性研究. 植物分类与资源学报, 35 (2):119-127. | |
| [5] | de Spiegelaere W, Dern-Wieloch J, Weigel R, Schumacher V, Schorle H, Nettersheim D, Bergmann M, Brehm R, Kliesch S, Vandekerckhove L, Fink C. 2015. Reference gene validation for RT-qPCR,a note on different available software packages. PLoS ONE, 10 (3):e122515. |
| [6] | Dong Lina, Liu Yan. 2019. Supplement to Begonia L. in Flora of Guangxi. Guihaia, 39 (1):16-39. (in Chinese) |
| 董莉娜, 刘演. 2019. 《广西植物志》秋海棠属(Begonia L.)增订. 广西植物, 39 (1):16-39. | |
| [7] |
Dong Y, Qu Y, Qi R, Bai X, Tian G, Wang Y, Wang J, Zhang K. 2018. Transcriptome analysis of the biosynthesis of anthocyanins in Begonia semperflorens under low-temperature and high-light conditions. Forests, 9 (2):87.
doi: 10.3390/f9020087 URL |
| [8] |
Fu Z Z, Shang H Q, Jiang H, Gao J, Dong X Y, Wang H J, Li Y M, Wang L M, Zhang J, Shu Q Y, Chao Y C, Xu M L, Wang R, Wang L S, Zhang H C. 2020. Systematic identification of the light-quality responding anthocyanin synthesis-related transcripts in petunia petals. Horticultural Plant Journal, 6 (6):428-438.
doi: 10.1016/j.hpj.2020.11.006 URL |
| [9] |
Gutierrez L, Mauriat M, Guenin S, Pelloux J, Lefebvre J F, Louvet R, Rusterucci C, Moritz T, Guerineau F, Bellini C, van Wuytswinkel O. 2008. The lack of a systematic validation of reference genes:a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR)analysis in plants. Plant Biotechnol J, 6 (6):609-618.
doi: 10.1111/j.1467-7652.2008.00346.x pmid: 18433420 |
| [10] |
Harborne J B, Williams C A. 2000. Advances in flavonoid research since 1992. Phytochemistry, 55 (6):481-504.
pmid: 11130659 |
| [11] |
Holton T A, Cornish E C. 1995. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell, 7 (7):1071-1083.
doi: 10.2307/3870058 URL |
| [12] |
Hughes N M, Vogelmann T C, Smith W K. 2008. Optical effects of abaxial anthocyanin on absorption of red wavelengths by understorey species:revisiting the back-scatter hypothesis. J Exp Bot, 59 (12):3435-3442.
doi: 10.1093/jxb/ern193 URL |
| [13] |
Khaldoun O A S, Abd E H. 2015. Biochemical and genetic evidences of anthocyanin biosynthesis and accumulation in a selected tomato mutant. Rendiconti Lincei, 26 (3):293-306.
doi: 10.1007/s12210-015-0446-x URL |
| [14] |
Li Y M, Zhang K M, Jin H H, Zhu L, Li Y H. 2015. Isolation and expression analysis of four putative structural genes involved in anthocyanin biosynthesis in Begonia semperflorens. J Hortic Sci Biotech, 90 (4):444-450.
doi: 10.1080/14620316.2015.11513208 URL |
| [15] |
Li Y, Qu Y, Wang Y, Bai X, Tian G, Liu Z, Li Y, Zhang K. 2019. Selection of suitable reference genes for qRT-PCR analysis of Begonia semperflorens under stress conditions. Mol Biol Rep, 46 (6):6027-6037.
doi: 10.1007/s11033-019-05038-5 URL |
| [16] |
Li W X, Wang L, He Z C, Lu Z G, Cui J W, Xu N T, Jin B, Wang L. 2020. Physiological and transcriptomic changes during autumn coloration and senescence in Ginkgo biloba leaves. Horticultural Plant Journal, 6 (6):396-408.
doi: 10.1016/j.hpj.2020.11.002 URL |
| [17] | Liang Lijun, Yang Yichen, Wang Erhuan, Xing Bingcong, Liang Zongsuo. 2018. Research progress on biosynthesis and regulation of plant anthocyanin. Journal of Anhui Agricultural Sciences, 46 (21):18-24. (in Chinese) |
| 梁立军, 杨祎辰, 王二欢, 邢丙聪, 梁宗锁. 2018. 植物花青素生物合成与调控研究进展. 安徽农业科学, 46 (21):18-24. | |
| [18] |
Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆C(T) method. Methods, 25 (4):402-408.
pmid: 11846609 |
| [19] |
Maroufi A, van Bockstaele E, de Loose M. 2010. Validation of reference genes for gene expression analysis in chicory(Cichorium intybus)using quantitative real-time PCR. BMC Mol Biol, 11:15.
doi: 10.1186/1471-2199-11-15 pmid: 20156357 |
| [20] | Mei Beijian, Ai Hua. 1987. Rapid propagation of iron cross Begonia. Plant Physiology Physiol Communications,(2):27-30. (in Chinese) |
| 梅贝坚, 艾华. 1987. 铁十字秋海棠试管快速繁殖. 植物生理学通讯,(2):27-30. | |
| [21] |
Pfaffl M W, Tichopad A, Prgomet C, Neuvians T P. 2004. Determination of stable housekeeping genes,differentially regulated target genes and sample integrity:BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol Lett, 26 (6):509-515.
doi: 10.1023/B:BILE.0000019559.84305.47 URL |
| [22] | Ren Yan, Zhang Weiguang. 2006. Research advances of anthocyanin. China Food Additives,(4):71-77. (in Chinese) |
| 任雁, 张惟广. 2006. 花色素苷的研究进展. 中国食品添加剂,(4):71-77. | |
| [23] |
Shi M Z, Xie D Y. 2014. Biosynthesis and metabolic engineering of anthocyanins in Arabidopsis thaliana. Recent Pat Biotechnol, 8 (1):47-60.
doi: 10.2174/1872208307666131218123538 URL |
| [24] |
Steyn W J, Wand S J E, Holcroft D M, Jacobs G. 2002. Anthocyanins in vegetative tissues:a proposed unified function in photoprotect. New Phytol, 155 (3):349-361.
doi: 10.1046/j.1469-8137.2002.00482.x pmid: 33873306 |
| [25] |
Sparvoli F, Martin C, Scienza A, Gavazzi G, Tonelli C. 1994. Cloning and molecular analysis of structural genes involved in flavonoid and stilbene biosynthesis in grape(Vitis vinifera L.). Plant Mol Biol, 24 (5):743-755.
pmid: 8193299 |
| [26] |
Tanaka Y, Sasaki N, Ohmiya A. 2008. Biosynthesis of plant pigments:anthocyanins,betalains and carotenoids. Plant J, 54 (4):733-749.
doi: 10.1111/j.1365-313X.2008.03447.x URL |
| [27] | Vandesompele J, de Preter K, Pattyn F, Poppe B, van Roy N, de Paepe A, Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol, 3 (7):H34. |
| [28] |
Wang J, Guo M, Li Y, Wu R, Zhang K. 2018. High-throughput transcriptome sequencing reveals the role of anthocyanin metabolism in Begonia semperflorens under high light stress. Photochem Photobiol, 94 (1):105-114.
doi: 10.1111/php.2018.94.issue-1 URL |
| [29] |
Winkel-Shirley B. 2001. Flavonoid biosynthesis. A colorful model for genetics,biochemistry,cell biology,and biotechnology. Plant Physiol, 126 (2):485-493.
pmid: 11402179 |
| [30] |
Wu Z J, Tian C, Jiang Q, Li X H, Zhuang J. 2016. Selection of suitable reference genes for qRT-PCR normalization during leaf development and hormonal stimuli in tea plant(Camellia sinensis). Sci Rep, 6:19748.
doi: 10.1038/srep19748 URL |
| [31] | Zeng Defu, Zhou Jianchan, Zhong Chunmei, Xie Jun. 2018. Screening of reference genes in Dioscorea composita tubers of different development stages. Plant Physiology Journal, 54 (3):509-517. (in Chinese) |
| 曾德福, 周建婵, 钟春梅, 谢君. 2018. 菊叶薯蓣不同发育时期块茎内参基因的筛选. 植物生理学报, 54 (3):509-517. | |
| [32] |
Zhang K M, Wang J W, Guo M L, Du W L, Wu R H, Wang X. 2016. Short-day signals are crucial for the induction of anthocyanin biosynthesis in Begonia semperflorens under low temperature condition. J Plant Physiol, 204:1-7.
doi: 10.1016/j.jplph.2016.06.021 URL |
| [33] | Zhang Ning, Hu Zongli, Chen Xuqing, Hou Xiaoshu, Li Yong, Chen Guoping. 2008. Analysis of metabolic pathway and establishment of regulating model of anthocyanin synthesis. China Biotechnology, 28 (1):97-105.. (in Chinese) |
| 张宁, 胡宗利, 陈绪清, 侯晓姝, 李勇, 陈国平. 2008. 植物花青素代谢途径分析及调控模型建立. 中国生物工程杂志, 28 (1):97-105. |
| [1] | 赵雪艳, 王琪, 王莉, 王方圆, 王庆, 李艳. 基于比较转录组的延胡索组织差异性表达分析[J]. 园艺学报, 2023, 50(1): 177-187. |
| [2] | 李镇希, 潘睿翾, 许美容, 郑正, 邓晓玲. 柑橘黄龙病菌双重实时荧光PCR检测方法的建立[J]. 园艺学报, 2023, 50(1): 188-196. |
| [3] | 高彦龙, 吴玉霞, 张仲兴, 王双成, 张瑞, 张德, 王延秀. 苹果ELO家族基因鉴定及其在低温胁迫下的表达分析[J]. 园艺学报, 2022, 49(8): 1621-1636. |
| [4] | 邱子文, 刘林敏, 林永盛, 林晓洁, 李永裕, 吴少华, 杨超. 千层金MbEGS基因的克隆与功能分析[J]. 园艺学报, 2022, 49(8): 1747-1760. |
| [5] | 郑林, 王帅, 刘语诺, 杜美霞, 彭爱红, 何永睿, 陈善春, 邹修平. 柑橘响应黄龙病菌侵染的NAC基因的克隆及表达分析[J]. 园艺学报, 2022, 49(7): 1441-1457. |
| [6] | 张秋悦, 刘昌来, 于晓晶, 杨甲定, 封超年. 盐胁迫条件下杜梨叶片差异表达基因qRT-PCR内参基因筛选[J]. 园艺学报, 2022, 49(7): 1557-1570. |
| [7] | 马维峰, 李艳梅, 马宗桓, 陈佰鸿, 毛娟. 苹果POD家族基因的鉴定与MdPOD15的功能分析[J]. 园艺学报, 2022, 49(6): 1181-1199. |
| [8] | 张凯, 麻明英, 王萍, 李益, 金燕, 盛玲, 邓子牛, 马先锋. 柑橘HSP20家族基因鉴定及其响应溃疡病菌侵染表达分析[J]. 园艺学报, 2022, 49(6): 1213-1232. |
| [9] | 梁晨, 孙如意, 向锐, 孙艺萌, 师校欣, 杜国强, 王莉. 葡萄生长调控因子GRF家族基因的鉴定及表达分析[J]. 园艺学报, 2022, 49(5): 995-1007. |
| [10] | 肖学宸, 刘梦雨, 蒋梦琦, 陈燕, 薛晓东, 周承哲, 吴兴健, 吴君楠, 郭寅生, 叶开温, 赖钟雄, 林玉玲. 龙眼褪黑素合成途径SNAT、ASMT和COMT家族基因鉴定及表达分析[J]. 园艺学报, 2022, 49(5): 1031-1046. |
| [11] | 高玮林, 张力曼, 薛超玲, 张垚, 刘孟军, 赵锦. 枣E类MADS基因在花和果中的表达及其蛋白互作研究[J]. 园艺学报, 2022, 49(4): 739-748. |
| [12] | 刘梦雨, 蒋梦琦, 陈燕, 张舒婷, 薛晓东, 肖学宸, 赖钟雄, 林玉玲. 龙眼GDSL酯酶/脂肪酶基因的全基因组鉴定及表达分析[J]. 园艺学报, 2022, 49(3): 597-612. |
| [13] | 姜翠翠, 方智振, 周丹蓉, 林炎娟, 叶新福. ‘芙蓉李’糖转运蛋白家族基因鉴定及表达分析[J]. 园艺学报, 2022, 49(2): 252-264. |
| [14] | 王智宇, 常贝贝, 刘琦, 程晓帆, 杜晓云, 于晓丽, 宋来庆, 赵玲玲. 苹果溶质转运蛋白基因MdSLC35F2-like表达与花青苷积累的研究[J]. 园艺学报, 2022, 49(11): 2293-2303. |
| [15] | 黄仁维, 任迎虹, 祁伟亮, 曾睿, 刘欣宇, 邓彬艳. 桑树MaERF105-Like的克隆及其在干旱胁迫下的表达分析[J]. 园艺学报, 2022, 49(11): 2439-2448. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司