
园艺学报 ›› 2021, Vol. 48 ›› Issue (11): 2227-2238.doi: 10.16420/j.issn.0513-353x.2020-0952
收稿日期:2021-07-30
修回日期:2021-10-13
发布日期:2021-12-02
通讯作者:
王华森,徐云敏
E-mail:wanghs@zafu.edu.cn;xuyunmin@zafu.edu.cn
基金资助:
WANG Shuwen, YANG Aiyi, WANG Huasen( ), XU Yunmin(
), XU Yunmin( )
)
Received:2021-07-30
Revised:2021-10-13
Published:2021-12-02
Contact:
WANG Huasen,XU Yunmin
E-mail:wanghs@zafu.edu.cn;xuyunmin@zafu.edu.cn
摘要:
以黄瓜‘9930’为试验材料,统计不同株龄黄瓜植株表型,注释黄瓜miR156/157-SPL途径基因,对基因结构、分类和表达进行分析。结果表明,黄瓜具有株龄表型特征,1/2叶期为幼苗期,3/4叶期为过渡期,5叶期为成株期。黄瓜基因组中共有7个Csa-MIR156位点,3个Csa-MIR157位点和14个Csa-SPL基因,分别位于第1、2、3、4和6号染色体。Csa-SPL基因的外显子数2 ~ 10个不等,Csa-SPL蛋白的SBP结构域由第1和第2外显子共同编码,SBP结构域含2个锌指结构域(Zn_1和Zn_2)和1个核定位信号。大分子量Csa-SPL8和Csa-SPL11还具有额外蛋白结构域。亲缘关系分析将Csa-SPL归为5个亚家族,其中11个Csa-SPL为Csa-miR156/157的靶基因,Csa-SPL5、10和14的Csa-miR156/157识别位点位于基因的3′UTR。Csa-SPL7为Csa-miR156靶基因,但Csa-miR156识别位点第11位碱基存在变异,且该变异在葫芦科植物保守存在。基因表达检测显示Csa-MIR156B和C为主效位点,Csa-miR156表达量随株龄增加而下降,靶基因Csa-SPL表达量则随株龄增加而上升。特殊的是,Csa-SPL7在黄瓜子叶期高表达,且不随株龄变化而变化,Csa-SPL7的表达特征同Csa-miR156识别位点碱基变异相吻合,表明Csa-SPL7已解除Csa-miR156的负调控作用。研究结果提示miR156/157-SPL对黄瓜营养期表型(叶形、节间长度和卷须发生)具有调控作用;此外,葫芦科植物miR156-SPL7负调控作用的解除,可能在其营养期早期发育行使特殊功能。
中图分类号:
汪淑雯, 杨爱怡, 王华森, 徐云敏. 黄瓜miR156/157-SPL途径基因鉴定和表达分析[J]. 园艺学报, 2021, 48(11): 2227-2238.
WANG Shuwen, YANG Aiyi, WANG Huasen, XU Yunmin. Identification and Expression Analysis of miR156/157-SPL Pathway Genes in Cucumber[J]. Acta Horticulturae Sinica, 2021, 48(11): 2227-2238.
| 基因名称 Gene name | 正向引物(5′-3′) Forward primer | 反向引物(5′-3′) Reverse primer |
|---|---|---|
| Csa-SPL1 | CCCTGTCACGAAGTATTTTGTCTC | TGTTTATCAGGAAAGTGGGTTGG |
| Csa-SPL2 | GCAGCATCAACCGACCGACTATCA | ACCAAAGGACGACCAATGGGAAT |
| Csa-SPL3 | CCACCAAACTACCTTATCAAGCG | GAAAGCACGAGGAATGTGAACTA |
| Csa-SPL4 | TAACGCCTCCATTTCTTTGCTCC | TAGATGGGGATTGATTTCCAGGATT |
| Csa-SPL5 | GCGACTGGAAAAGCTGATGGAA | ATCCCCTGAAAACCCTAATTCTGC |
| Csa-SPL6 | GAAGAACGAATCATGGGCCAACT | TCCGAAATCATCCGTCCCTCC |
| Csa-SPL7 | CAGTCCCATCTGAGTGTTCCAGT | CTTCCTCCATTTCAAGACCCA |
| Csa-SPL8 | GAAGGAAGGAGTAATGCGATGGA | GAACCGTGGCAGTGAAAAGAG |
| Csa-SPL9 | TCCAGAAACCGAACGACGACT | GCATGTGAAACTTGGTGATTGAGAC |
| Csa-SPL10 | AGAGCCAAGGGGAAAGCACAA | CTTCCTGGAGGAGCCATCTGAAT |
| Csa-SPL11 | ATCACCTACTACTCTTGGTCGTTGT | CTTAGCCTTGGTTGCGAAGA |
| Csa-SPL12 | TATGTTGCCCGGTTCGTTGT | CCGTTTCCGAGCACTTTCTG |
| Csa-SPL13 | CTAACTCATCGCAACTGATTCAAAG | TGGTTAAGGCAGTCTAGTGACATTCT |
| Csa-SPL14 | CTCAGAAGATCAAGGTTGGGAGG | ACCACCCCGAGCACTGGTTA |
| Csa-MIR156A | GTATTGAAAATTAGAGTAAGGGGAAG | CTTCAAGCATGAACCCTAACAT |
| Csa-MIR156B | GAGAGAAAAGCACAAAAGACCAAGA | CTAAGTAAAGGTATGAACTTTCAACTT |
| 基因名称 Gene name | 正向引物(5′-3′) Forward primer | 反向引物(5′-3′) Reverse primer |
| Csa-MIR156C | GTTTATCGTTCTTGAATTTGGTTAG | ATACACTTTATAGCAACTGCCTCTG |
| Csa-MIR156D | TGAATGATGGTGAGTGTGTTAGGAG | CACGCACCCGCAAAGGTAT |
| Csa-MIR156E | AAAGGTGATTAAAATTGAGGGGAA | TGGCATATTATTATACCAAAAGCCT |
| Csa-MIR156F | CATACGGAAGGTAATCTCAAGGA | AATATCTAGCATAAGCGGCCATG |
| Csa-MIR156G | TCTCACGGTCTATTTCTAACACG | CACAAAGCTTCGAGCATTGATAT |
| Csa-MIR157A | GGGTTTAGAAATTTGGAGAGAGACA | TATTAAAAGTGAAGGGATGGGATATA |
| Csa-MIR157B | ATTTATCATGCACAAGGGAGAACTT | AATTGCTATTAGTGGTACCGATTGA |
| Csa-MIR157C | CCAATACGGTGATAGCTATATGTTGT | TAACAAAGTGGTGTTTACGACTCTA |
| Csa-Tub | CACTACACCGTTGGAAAGGAAA | CAAAAGGAGGGAGCCGAGA |
| Csa-U6 | GGGGACATCCGATAAAATT | TGTGCGTGTCATCCTTGC |
| Csa-miR156 | GCGGCGGTGACAGAAGAGAG | GTGCAGGGTCCGAGGT |
| Csa-miR156 RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCAC | TGGATACGACGTGCTC |
表1 基因表达检测引物
Table 1 Gene detection primers
| 基因名称 Gene name | 正向引物(5′-3′) Forward primer | 反向引物(5′-3′) Reverse primer |
|---|---|---|
| Csa-SPL1 | CCCTGTCACGAAGTATTTTGTCTC | TGTTTATCAGGAAAGTGGGTTGG |
| Csa-SPL2 | GCAGCATCAACCGACCGACTATCA | ACCAAAGGACGACCAATGGGAAT |
| Csa-SPL3 | CCACCAAACTACCTTATCAAGCG | GAAAGCACGAGGAATGTGAACTA |
| Csa-SPL4 | TAACGCCTCCATTTCTTTGCTCC | TAGATGGGGATTGATTTCCAGGATT |
| Csa-SPL5 | GCGACTGGAAAAGCTGATGGAA | ATCCCCTGAAAACCCTAATTCTGC |
| Csa-SPL6 | GAAGAACGAATCATGGGCCAACT | TCCGAAATCATCCGTCCCTCC |
| Csa-SPL7 | CAGTCCCATCTGAGTGTTCCAGT | CTTCCTCCATTTCAAGACCCA |
| Csa-SPL8 | GAAGGAAGGAGTAATGCGATGGA | GAACCGTGGCAGTGAAAAGAG |
| Csa-SPL9 | TCCAGAAACCGAACGACGACT | GCATGTGAAACTTGGTGATTGAGAC |
| Csa-SPL10 | AGAGCCAAGGGGAAAGCACAA | CTTCCTGGAGGAGCCATCTGAAT |
| Csa-SPL11 | ATCACCTACTACTCTTGGTCGTTGT | CTTAGCCTTGGTTGCGAAGA |
| Csa-SPL12 | TATGTTGCCCGGTTCGTTGT | CCGTTTCCGAGCACTTTCTG |
| Csa-SPL13 | CTAACTCATCGCAACTGATTCAAAG | TGGTTAAGGCAGTCTAGTGACATTCT |
| Csa-SPL14 | CTCAGAAGATCAAGGTTGGGAGG | ACCACCCCGAGCACTGGTTA |
| Csa-MIR156A | GTATTGAAAATTAGAGTAAGGGGAAG | CTTCAAGCATGAACCCTAACAT |
| Csa-MIR156B | GAGAGAAAAGCACAAAAGACCAAGA | CTAAGTAAAGGTATGAACTTTCAACTT |
| 基因名称 Gene name | 正向引物(5′-3′) Forward primer | 反向引物(5′-3′) Reverse primer |
| Csa-MIR156C | GTTTATCGTTCTTGAATTTGGTTAG | ATACACTTTATAGCAACTGCCTCTG |
| Csa-MIR156D | TGAATGATGGTGAGTGTGTTAGGAG | CACGCACCCGCAAAGGTAT |
| Csa-MIR156E | AAAGGTGATTAAAATTGAGGGGAA | TGGCATATTATTATACCAAAAGCCT |
| Csa-MIR156F | CATACGGAAGGTAATCTCAAGGA | AATATCTAGCATAAGCGGCCATG |
| Csa-MIR156G | TCTCACGGTCTATTTCTAACACG | CACAAAGCTTCGAGCATTGATAT |
| Csa-MIR157A | GGGTTTAGAAATTTGGAGAGAGACA | TATTAAAAGTGAAGGGATGGGATATA |
| Csa-MIR157B | ATTTATCATGCACAAGGGAGAACTT | AATTGCTATTAGTGGTACCGATTGA |
| Csa-MIR157C | CCAATACGGTGATAGCTATATGTTGT | TAACAAAGTGGTGTTTACGACTCTA |
| Csa-Tub | CACTACACCGTTGGAAAGGAAA | CAAAAGGAGGGAGCCGAGA |
| Csa-U6 | GGGGACATCCGATAAAATT | TGTGCGTGTCATCCTTGC |
| Csa-miR156 | GCGGCGGTGACAGAAGAGAG | GTGCAGGGTCCGAGGT |
| Csa-miR156 RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCAC | TGGATACGACGTGCTC |
| 基因名称 Gene name | 基因编号 Gene ID | 长度/bp Length | 染色体 Chromosome | 位置 Location |
|---|---|---|---|---|
| Csa-SPL1 | Csa1G001450 | 379 | Chr1 | 237765..239802 (+) |
| Csa-SPL2 | Csa1G015680 | 550 | Chr1 | 2088067..2093861 (+) |
| Csa-SPL3 | Csa1G039890 | 548 | Chr1 | 3934043..3940006 (-) |
| Csa-SPL4 | Csa1G051590 | 314 | Chr1 | 6001183..6004194 (-) |
| Csa-SPL5 | Csa1G074980 | 162 | Chr1 | 7700146..7703682 (+) |
| Csa-SPL6 | Csa3G117960 | 340 | Chr3 | 6445761..6448246 (+) |
| Csa-SPL7 | Csa3G567830 | 344 | Chr3 | 22275377..22277440 (+) |
| Csa-SPL8 | Csa3G664550 | 1 013 | Chr3 | 25953081..25964518 (+) |
| Csa-SPL9 | Csa3G809420 | 382 | Chr3 | 30985575..30991777 (+) |
| Csa-SPL10 | Csa4G631590 | 202 | Chr4 | 20657444..20658877 (-) |
| Csa-SPL11 | Csa4G664590 | 1 031 | Chr4 | 23212471..23218465 (+) |
| Csa-SPL12 | Csa6G094760 | 328 | Chr6 | 6509944..6513210 (+) |
| Csa-SPL13 | Csa6G109120 | 297 | Chr6 | 7313479..7317016 (-) |
| Csa-SPL14 | Csa6G517960 | 141 | Chr6 | 27284755..27286747 (+) |
| Csa-MIR156A | 127 | Chr1 | 13072567..13072694 (+) | |
| Csa-MIR156B | 112 | Chr2 | 1701701..1701813 (+) | |
| Csa-MIR156C | 121 | Chr2 | 19844384..19844505 (+) | |
| Csa-MIR156D | 175 | Chr4 | 22021976..22022151 (+) | |
| Csa-MIR156E | 138 | Chr4 | 22620675..22620813 (-) | |
| Csa-MIR156F | 89 | Chr1 | 22345006..22345095 (+) | |
| Csa-MIR156G | 148 | Chr6 | 16464371..16464519 (-) | |
| Csa-MIR157A | 165 | Chr1 | 2522433..2522598 (+) | |
| Csa-MIR157B | 177 | Chr3 | 22790035..22790212 (-) | |
| Csa-MIR157C | 85 | Chr2 | 20893540..20893625 (-) |
表2 黄瓜miR156/157-SPL途径基因信息
Table 2 The information of miR156/157-SPL pathway genes in cucumber
| 基因名称 Gene name | 基因编号 Gene ID | 长度/bp Length | 染色体 Chromosome | 位置 Location |
|---|---|---|---|---|
| Csa-SPL1 | Csa1G001450 | 379 | Chr1 | 237765..239802 (+) |
| Csa-SPL2 | Csa1G015680 | 550 | Chr1 | 2088067..2093861 (+) |
| Csa-SPL3 | Csa1G039890 | 548 | Chr1 | 3934043..3940006 (-) |
| Csa-SPL4 | Csa1G051590 | 314 | Chr1 | 6001183..6004194 (-) |
| Csa-SPL5 | Csa1G074980 | 162 | Chr1 | 7700146..7703682 (+) |
| Csa-SPL6 | Csa3G117960 | 340 | Chr3 | 6445761..6448246 (+) |
| Csa-SPL7 | Csa3G567830 | 344 | Chr3 | 22275377..22277440 (+) |
| Csa-SPL8 | Csa3G664550 | 1 013 | Chr3 | 25953081..25964518 (+) |
| Csa-SPL9 | Csa3G809420 | 382 | Chr3 | 30985575..30991777 (+) |
| Csa-SPL10 | Csa4G631590 | 202 | Chr4 | 20657444..20658877 (-) |
| Csa-SPL11 | Csa4G664590 | 1 031 | Chr4 | 23212471..23218465 (+) |
| Csa-SPL12 | Csa6G094760 | 328 | Chr6 | 6509944..6513210 (+) |
| Csa-SPL13 | Csa6G109120 | 297 | Chr6 | 7313479..7317016 (-) |
| Csa-SPL14 | Csa6G517960 | 141 | Chr6 | 27284755..27286747 (+) |
| Csa-MIR156A | 127 | Chr1 | 13072567..13072694 (+) | |
| Csa-MIR156B | 112 | Chr2 | 1701701..1701813 (+) | |
| Csa-MIR156C | 121 | Chr2 | 19844384..19844505 (+) | |
| Csa-MIR156D | 175 | Chr4 | 22021976..22022151 (+) | |
| Csa-MIR156E | 138 | Chr4 | 22620675..22620813 (-) | |
| Csa-MIR156F | 89 | Chr1 | 22345006..22345095 (+) | |
| Csa-MIR156G | 148 | Chr6 | 16464371..16464519 (-) | |
| Csa-MIR157A | 165 | Chr1 | 2522433..2522598 (+) | |
| Csa-MIR157B | 177 | Chr3 | 22790035..22790212 (-) | |
| Csa-MIR157C | 85 | Chr2 | 20893540..20893625 (-) |
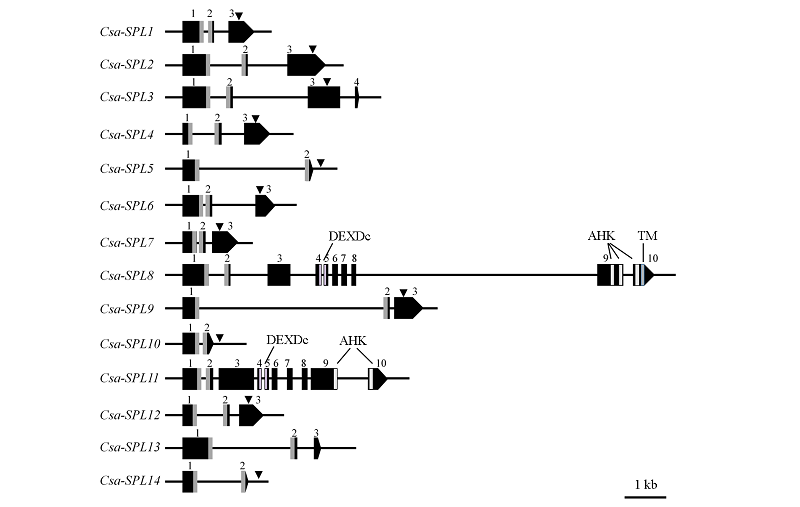
图3 黄瓜Csa-SPL基因结构分析结果 数字:外显子;灰色:SBP结构域的编码区;倒三角形:miR156识别位点。
Fig. 3 The gene structure of Csa-SPL in cucumber Number:Exon. Gray:SBP domain coding region. Inverted triangle:miR156 binding site.
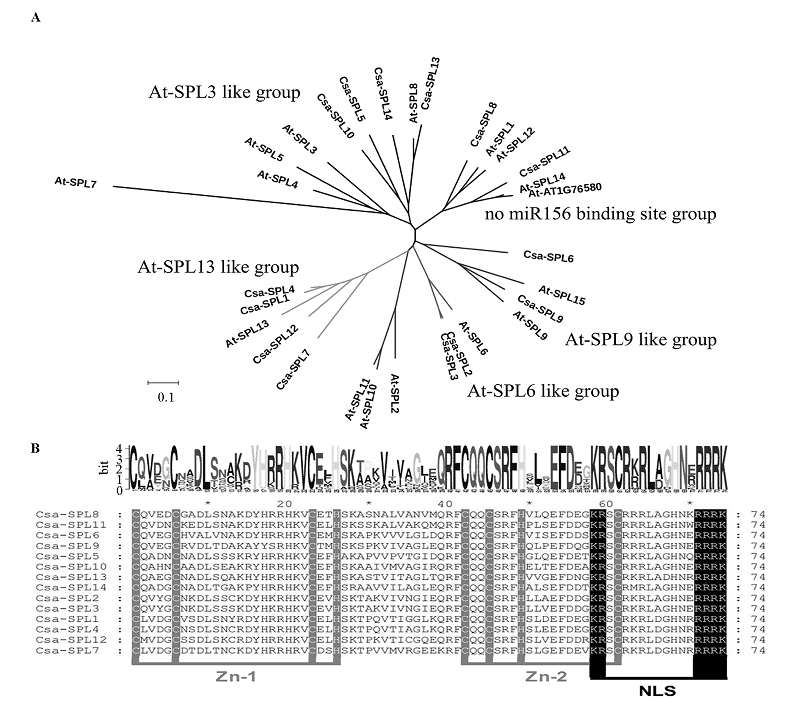
图4 黄瓜Csa-SPL和拟南芥At-SPL蛋白聚类(A)和Csa-SBP结构域(B)分析 Zn_1和Zn_2表示锌指结构域,NLS表示核定位信号序列。
Fig. 4 NJ tree analyzing(A)and alignment of SBP domain(B)results of Csa-SPL protein sequences Zn_1 and Zn_2 indicate the Zinc finger domains,NLS indicates the nuclear localization signal.
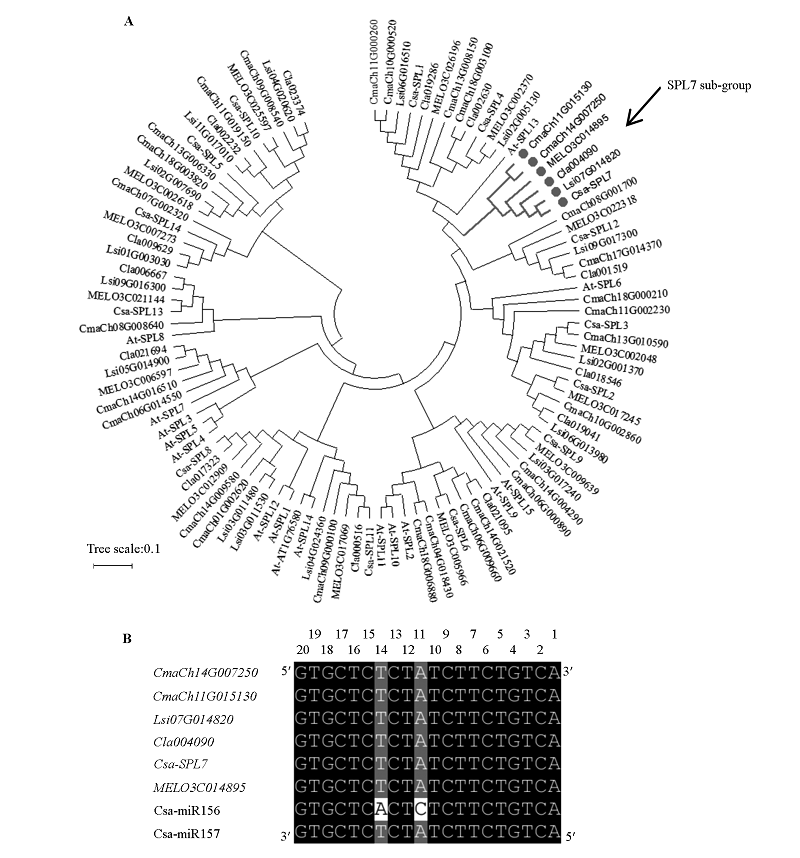
图6 葫芦科植物SPL聚类(A)和SPL7的miR156/157识别位点序列比对(B)结果
Fig. 6 NJ tree analyzing of Cucurbitaceae SPL(A)and alignment of miR156/157 binding sites of Cucurbitaceae SPL7(B)
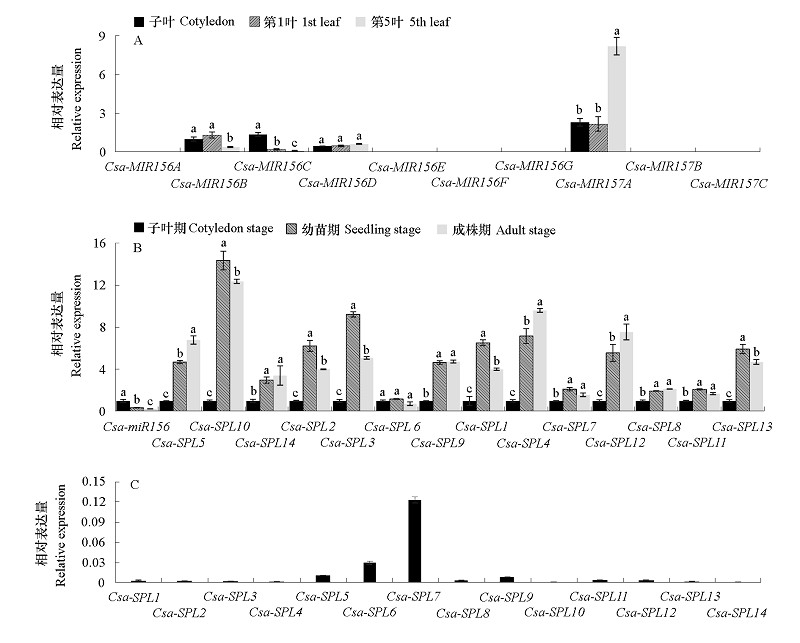
图7 黄瓜miR156/157-SPL途径基因表达检测结果 A:不同叶片Csa-MIR156/157检测结果;B:不同时期茎尖Csa-miR156和Csa-SPL检测结果(各基因在子叶期的表达量归一为1);C:子叶期茎尖各Csa-SPL检测结果。不同小写字母表示0.05水平差异显著。
Fig. 7 Detection of miR156/157-SPLs pathway genes expression in cucumber A. Expression of Csa-MIR156/157 in different leaves;B. Expression of Csa-miR156 and Csa-SPL in different stages(The gene expression levels in cotyledon stage are normalized to 1);C. Expression of Csa-SPLs in cotyledon stage. Different letters represent significant differences at the level of 0.05.
| [1] |
Birkenbihl R P, Jach G, Saedler H, Huijser P. 2005. Functional dissection of the plant-specific SBP-domain:overlap of the DNA-binding and nuclear localization domains. Journal of Molecular Biology, 352 (3):585-596.
pmid: 16095614 |
| [2] |
Cui L, Zheng F, Wang J, Zhang C, Xiao F, Ye J, Li C, Ye Z, Zhang J. 2020. miR156a-targeted SBP-Box transcription factor SlSPL 13 regulates inflorescence morphogenesis by directly activating SFT in tomato. Plant Biotechnology Journal, 18 (8):1670-1682.
doi: 10.1111/pbi.v18.8 URL |
| [3] |
Gou J, Felippes F F, Liu C, Weigel D, Wang J. 2011. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription Factor. The Plant Cell, 23 (4):1512-1522.
doi: 10.1105/tpc.111.084525 URL |
| [4] |
He J, Xu M, Willmann M R, McCormick K, Hu T, Yang L, Starker C G, Voytas D F, Meyers B C, Poethig R S. 2018. Threshold-dependent repression of SPL gene expression by miR156/miR157 controls vegetative phase change in Arabidopsis thaliana. PLoS Genetics, 14 (4):e1007337.
doi: 10.1371/journal.pgen.1007337 URL |
| [5] |
Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, Dong G, Zeng D, Lu Z, Zhu X, Qian Q, Li J. 2010. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nature Genetics, 42 (6):541-544.
doi: 10.1038/ng.591 URL |
| [6] | Li Meng-ting. 2017. Cloning and preliminary function analysis of homologous SPL genes in cucumber(Cucumis sativus L.)[M. D. Dissertation]. Hangzhou: Hangzhou Normal University. (in Chinese) |
| 李梦婷. 2017. 黄瓜SPL同源基因的克隆及功能的初步研究[硕士论文]. 杭州: 杭州师范大学. | |
| [7] |
Liu Q, Harberd N P, Fu X. 2016a. SQUAMOSA promoter binding protein-like transcription factors: targets for improving cereal grain yield. Molecular Plant, 9 (6):765-767.
doi: 10.1016/j.molp.2016.04.008 URL |
| [8] | Liu R, Lai B, Hu B, Qin Y, Hu G, Zhao J. 2016b. Identification of MicroRNAs and their target genes related to the accumulation of anthocyanins in Litchi chinensis by high-throughput sequencing and degradome analysis. Frontiers in Plant Science, 7:2059. |
| [9] |
Luo Y, Guo Z, Li L. 2013. Evolutionary conservation of microRNA regulatory programs in plant flower development. Development Biology, 380 (2):133-144.
doi: 10.1016/j.ydbio.2013.05.009 URL |
| [10] |
Ma Y, Xue H, Zhang F, Jiang Q, Yang S, Yue P, Wang F, Zhang Y, Li L, He P, Zhang Z. 2020. The miR156/SPL module regulates apple salt stress tolerance by activating MdWRKY100 expression. Plant Biotechnology Journal, 19 (2):311-323.
doi: 10.1111/pbi.v19.2 URL |
| [11] |
Mao W, Li Z, Xia X, Li Y, Yu J. 2012. A combined approach of high-throughput sequencing and degradome analysis reveals tissue specific expression of microRNAs and their targets in cucumber. PLoS ONE, 7 (3):e33040.
doi: 10.1371/journal.pone.0033040 URL |
| [12] |
Martinez G, Forment J, Llave C, Pallas V, Gomez G. 2011. High-throughput sequencing, characterization and detection of new and conserved cucumber miRNAs. PLoS ONE, 6 (5):e19523.
doi: 10.1371/journal.pone.0019523 URL |
| [13] |
Rubio-Somoza I, Zhou C, Confraria A, Martinho C, von Born P, Baena-Gonzalez E, Wang J, Weigel D. 2014. Temporal control of leaf complexity by miRNA-regulated licensing of protein complexes. Current Biology, 24 (22):2714-2719.
doi: 10.1016/j.cub.2014.09.058 pmid: 25448000 |
| [14] |
Schwab R, Palatnik J F, Riester M, Schommer C, Schmid M, Weigel D. 2005. Specific effects of microRNAs on the plant transcriptome. Developmental Cell, 8 (4):517-527.
doi: 10.1016/j.devcel.2005.01.018 URL |
| [15] |
Silva G F F E, Silva E M, Da Silva Azevedo M, Guivin M A C, Ramiro D A, Figueiredo C R, Carrer H, Peres L E P, Nogueira F T S. 2014. microRNA156-targeted SPL/SBP box transcription factors regulate tomato ovary and fruit development. The Plant Journal, 78 (4):604-618.
doi: 10.1111/tpj.12493 URL |
| [16] |
Wang J, Czech B, Weigel D. 2009. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell, 138 (4):738-749.
doi: 10.1016/j.cell.2009.06.014 URL |
| [17] |
Wang J, Zhou L, Shi H, Chern M, Yu H, Yi H, He M, Yin J, Zhu X, Li Y, Li W, Liu J, Wang J, Chen X, Qing H, Wang Y, Liu G, Wang W, Li P, Wu X, Zhu L, Zhou J M, Ronald P C, Li S, Li J, Chen X. 2018. A single transcription factor promotes both yield and immunity in rice. Science, 361 (6406):1026-1028.
doi: 10.1126/science.aat7675 URL |
| [18] | Wang L, Zhou C M, Mai Y X, Li L Z, Gao J, Shang G D, Lian H, Han L, Zhang T Q, Tang H B, Ren H, Wang F X, Wu L Y, Liu X L, Wang C S, Chen E W, Zhang X N, Liu C, Wang J W. 2019. A spatiotemporally regulated transcriptional complex underlies heteroblastic development of leaf hairs in Arabidopsis thaliana. EMBO Journal, 38 (8):e100063. |
| [19] |
Wang Y, Wu F, Bai J, He Y. 2014. BrpSPL9(Brassica rapa ssp. pekinensis SPL9)controls the earliness of heading time in Chinese cabbage. Plant Biotechnology Journal, 12 (3):312-321.
doi: 10.1111/pbi.2014.12.issue-3 URL |
| [20] |
Willmann M R, Poethig R S. 2007. Conservation and evolution of miRNA regulatory programs in plant development. Current Opinion in Plant Biology, 10 (5):503-511.
pmid: 17709279 |
| [21] |
Wu G, Park M Y, Conway S R, Wang J, Weigel D, Poethig R S. 2009. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell, 138 (4):750-759.
doi: 10.1016/j.cell.2009.06.031 URL |
| [22] |
Wu G, Poethig R S. 2006. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development, 133 (18):3539-3547.
doi: 10.1242/dev.02521 URL |
| [23] | Xu M, Hu T, Zhao J, Park M, Earley K W, Wu G, Yang L, Poethig R S. 2016. Developmental Functions of miR156-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLoS Genetics, 12 (8):e1006263. |
| [24] |
Xu Y, Qian Z, Zhou B, Wu G. 2019. Age-dependent heteroblastic development of leaf hairs in Arabidopsis. New Phytologist, 224 (2):741-748.
doi: 10.1111/nph.v224.2 URL |
| [25] |
Yamasaki K, Kigawa T, Inoue M, Tateno M, Yamasaki T, Yabuki T, Aoki M, Seki E, Matsuda T, Nunokawa E, Ishizuka Y, Terada T, Shirouzu M, Osanai T, Tanaka A, Seki M, Shinozaki K, Yokoyama S. 2004. A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors. Journal of Molecular Biology, 337 (1):49-63.
doi: 10.1016/j.jmb.2004.01.015 URL |
| [26] |
Yu N, Niu Q, Ng K, Chua N. 2015. The role of miR156/SPLs modules in Arabidopsis lateral root development. The Plant Journal, 83 (4):673-685.
doi: 10.1111/tpj.12919 pmid: 26096676 |
| [27] |
Zhang H, Zhang L, Han J, Qian Z, Zhou B, Xu Y, Wu G. 2019. The nuclear localization signal is required for the function of squamosa promoter binding protein-like gene 9 to promote vegetative phase change in Arabidopsis. Plant Molecular Biology, 100 (6):571-578.
doi: 10.1007/s11103-019-00863-5 pmid: 30953277 |
| [28] |
Zhang X, Zou Z, Zhang J, Zhang Y, Han Q, Hu T, Xu X, Liu H, Li H, Ye Z. 2011. Over-expression of sly-miR156a in tomato results in multiple vegetative and reproductive trait alterations and partial phenocopy of the sft mutant. FEBS Letters, 585 (2):435-439.
doi: 10.1016/j.febslet.2010.12.036 URL |
| [29] |
Zheng C, Ye M, Sang M, Wu R. 2019. A regulatory network for miR156-SPL module in Arabidopsis thaliana. International Journal of Molecular Science, 20 (24):6166.
doi: 10.3390/ijms20246166 URL |
| [1] | 王晓晨, 聂子页, 刘先菊, 段 伟, 范培格, 梁振昌, . 脱落酸对‘京香玉’葡萄果实单萜物质合成的影响[J]. 园艺学报, 2023, 50(2): 237-249. |
| [2] | 翟含含, 翟宇杰, 田义, 张叶, 杨丽, 温陟良, 陈海江. 桃SAUR家族基因分析及PpSAUR5功能鉴定[J]. 园艺学报, 2023, 50(1): 1-14. |
| [3] | 罗天宽, 吴海涛, 张圣美, 黄宗安, 孙 继, 水德聚, 陈先知. 黄瓜新品种‘瓯翠1号’[J]. 园艺学报, 2022, 49(S2): 125-126. |
| [4] | 王鹤冰, 向华丰, 陈新中, 张 生, 张洪成. 华南型黄瓜新品种‘新燕095’[J]. 园艺学报, 2022, 49(S1): 79-80. |
| [5] | 许春梅, 张作标, 柳景兰, 王 昕, 杨 龙, 赵 丹, 刘思宇, 贾云鹤, 孟雪娇, 崔嵩岑. 黄瓜新品种‘绿春2号’[J]. 园艺学报, 2022, 49(S1): 81-82. |
| [6] | 张利东, 黄洪宇, 孔维良, 李加旺, 李愚鹤, . 华北型黄瓜新品种‘津优355’[J]. 园艺学报, 2022, 49(S1): 83-84. |
| [7] | 王惠哲, 杨瑞环, 邓 强, 曹明明, 李淑菊, . 抗黑星病黄瓜新品种‘津冬369’[J]. 园艺学报, 2022, 49(S1): 85-86. |
| [8] | 聂鑫淼, 栾恒, 冯改利, 王超, 李岩, 魏珉. 硅营养和嫁接砧木对黄瓜幼苗耐冷性的影响[J]. 园艺学报, 2022, 49(8): 1795-1804. |
| [9] | 张秋悦, 刘昌来, 于晓晶, 杨甲定, 封超年. 盐胁迫条件下杜梨叶片差异表达基因qRT-PCR内参基因筛选[J]. 园艺学报, 2022, 49(7): 1557-1570. |
| [10] | 韩鲁杰, 冯一清, 杨秀华, 张宁, 毕焕改, 艾希珍. 有机肥化肥配施对大棚黄瓜根区土壤与根系特征的影响[J]. 园艺学报, 2022, 49(5): 1047-1059. |
| [11] | 李亚梅, 马福利, 张山奇, 黄锦秋, 陈梦婷, 周军永, 孙其宝, 孙俊. 酸枣愈伤组织转化体系构建及在ZjBRC1调控ZjYUCCA表达中的应用[J]. 园艺学报, 2022, 49(4): 749-757. |
| [12] | 权建华, 段誉, 罗天, 袁强, 齐鑫, 王勤礼. 黄瓜新品种‘裕研9号’[J]. 园艺学报, 2022, 49(3): 703-704. |
| [13] | 张瑞, 张夏燚, 赵婷, 王双成, 张仲兴, 刘博, 张德, 王延秀. 基于转录组分析垂丝海棠响应盐碱胁迫的分子机制[J]. 园艺学报, 2022, 49(2): 237-251. |
| [14] | 周至铭, 杨佳宝, 张程, 曾令露, 孟晚秋, 孙黎. 向日葵LACS家族鉴定及响应非生物胁迫表达分析[J]. 园艺学报, 2022, 49(2): 352-364. |
| [15] | 宋蒙飞, 查高辉, 陈劲枫, 娄群峰. 黄瓜株型性状分子基础研究进展[J]. 园艺学报, 2022, 49(12): 2683-2702. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司