
园艺学报 ›› 2021, Vol. 48 ›› Issue (10): 1847-1858.doi: 10.16420/j.issn.0513-353x.2021-0090
刘玥1, 李月庆2, 孟祥宇3, 黄靖1, 汤昊1, 李源恒1, 高翔2, 赵春莉1,*( )
)
收稿日期:2021-06-09
修回日期:2021-08-30
出版日期:2021-10-25
发布日期:2021-11-01
通讯作者:
赵春莉
E-mail:zcl8368@163.com
基金资助:
LIU Yue1, LI Yueqing2, MENG Xiangyu3, HUANG Jing1, TANG Hao1, LI Yuanheng1, GAO Xiang2, ZHAO Chunli1,*( )
)
Received:2021-06-09
Revised:2021-08-30
Online:2021-10-25
Published:2021-11-01
Contact:
ZHAO Chunli
E-mail:zcl8368@163.com
摘要:
为研究大花君子兰(Clivia miniata)查尔酮合酶的结构及功能,基于前期构建的大花君子兰转录组数据库,对参与花青素合成的CHS基因家族进行鉴定和筛选。在大花君子兰中克隆得到4个CmCHS(CmCHS1 ~ CmCHS4),其开放阅读框全长分别为1 173、1 170、1 173和1 173 bp,编码390、389、390、390个氨基酸。氨基酸序列比对分析证实,CmCHS具备该基因家族典型的保守结构域。进化树分析结果显示,4条CmCHS属于真实的CHS,编码蛋白属于Ⅲ型聚酮合成酶(PKS),亚细胞定位于细胞质和细胞核中。基因表达特异性分析结果表明,CmCHS1和CmCHS2在花朵初开和盛开期表达量较高;CmCHS1、CmCHS2、CmCHS3在红色叶片和果皮中的表达量大大高于绿色叶片和果皮,CmCHS4则相对较低。构建原核表达载体pET-32-CmCHS,体外酶活检测表明,4个CmCHS重组蛋白皆可催化香豆酰辅酶A和丙二酰辅酶A生成柚皮素查尔酮。
中图分类号:
刘玥, 李月庆, 孟祥宇, 黄靖, 汤昊, 李源恒, 高翔, 赵春莉. 大花君子兰查尔酮合酶基因CmCHS的克隆及其功能验证[J]. 园艺学报, 2021, 48(10): 1847-1858.
LIU Yue, LI Yueqing, MENG Xiangyu, HUANG Jing, TANG Hao, LI Yuanheng, GAO Xiang, ZHAO Chunli. Cloning and Functional Characterization of Chalcone Synthase Genes(CmCHS)from Clivia miniata[J]. Acta Horticulturae Sinica, 2021, 48(10): 1847-1858.
| 用途 Use | 基因 Gene | 引物序列(5′-3′) Primer sequence |
|---|---|---|
| 基因ORF的克隆 Cloning the full-length of ORF | CmCHS1 | F:ATGGTGAGTATCGATGAGATT;R:CATCGCGCCGATGATTAATT |
| CmCHS2 | F:ATGGTGAGTGTCCATGAGATTCGC;R:CACGATGTGTCGCATCGATAAATT | |
| CmCHS3 | F:ATGGTGAGCGTGGAAGAGATC;R:GGTATCACACCGATGATTAATT | |
| CmCHS4 | F:ATGGCCACAATTGAGGAGATCAG;R:ACGCTTCACACCGCTAGTTAACT | |
| T7 | F:TAATACGACTCACTATAGGG;R:GCTAGTTATTGCTCAGCGG | |
| 实时荧光定PCR Quantitative Real-time PCR | CmCHS1 | F:GACATGCCCGGTGCAGACTACCA;R:AGCCGTGATCTCCGAGCAGACG |
| CmCHS2 | F:GACGAGACAAGTGTTGAGCGAGTA;R:TCCAGGTCCGAATCCAAAGAGC | |
| CmCHS3 | F:AGGAGCGATAGACGGGCACTTA;R:GAACAGCGAGTTCCAATCTAACATC | |
| CmCHS4 | F:GGAGGTGCTGAAGGAGTATGGGAACA;R:TCCAGGGCCGAACCCAAATAGA | |
| CmActin | F:GCATCACACCTTCTACAA;R:CATTGTAGAAGGTGTGATG | |
| 制备重组蛋白 Preparation of recombinant protein | CmCHS1 | F:TGGCTGATATCGGATCCATGGTGAGTATCGATGAG; R:CGACGGAGCTCGAATTCTTAATTAGTAGCCGCGCT |
| CmCHS2 | F:TGGCTGATATCGGATCCATGGTGAGTGTCCATGAG; R:CGACGGAGCTCGAATTCTTAAATAGCTACGCTGTG | |
| CmCHS3 | F:TGGCTGATATCGGATCCATGGTGAGCGTGGAAGAG; R:CGACGGAGCTCGAATTCTTAATTAGTAGCCACACT | |
| CmCHS4 | F:TGGCTGATATCGGATCCATGGCCACAATTGAGGAG; R:CGACGGAGCTCGAATTCTCAATTGATCGCCACACT | |
| 亚细胞定位 Subcellular | GFP(35S:GFP) | F:CTGATTACGCTCATATGATGGTGAGCAAGGGCGAG; R:AGGATTCAATCTTAAGTTACTTGTACAGCTCGTCCATG |
| localization | (GFP)CmCHS1 | F:GACGAGCTGTACAAGCATATGGTGAGTATCGAT; R:CAACAGGATTCAATCTTAAGTTAATTAGTAGCCGC |
| (GFP)CmCHS2 | F:GACGAGCTGTACAAGCATATGGTGAGTGTCCAT; R:CAACAGGATTCAATCTTAAGTTAAATAGCTACGCT | |
| (GFP)CmCHS3 | F:GACGAGCTGTACAAGCATATGGTGAGCGTGGAA; R:CAACAGGATTCAATCTTAAGTTAATTAGTAGCCAC | |
| (GFP)CmCHS4 | F:GACGAGCTGTACAAGCATATGGCCACAATTGAG; R:CAACAGGATTCAATCTTAAGTCAATTGATCGCCAC |
表1 本研究中的引物序列
Table 1 Primer sequences in this study
| 用途 Use | 基因 Gene | 引物序列(5′-3′) Primer sequence |
|---|---|---|
| 基因ORF的克隆 Cloning the full-length of ORF | CmCHS1 | F:ATGGTGAGTATCGATGAGATT;R:CATCGCGCCGATGATTAATT |
| CmCHS2 | F:ATGGTGAGTGTCCATGAGATTCGC;R:CACGATGTGTCGCATCGATAAATT | |
| CmCHS3 | F:ATGGTGAGCGTGGAAGAGATC;R:GGTATCACACCGATGATTAATT | |
| CmCHS4 | F:ATGGCCACAATTGAGGAGATCAG;R:ACGCTTCACACCGCTAGTTAACT | |
| T7 | F:TAATACGACTCACTATAGGG;R:GCTAGTTATTGCTCAGCGG | |
| 实时荧光定PCR Quantitative Real-time PCR | CmCHS1 | F:GACATGCCCGGTGCAGACTACCA;R:AGCCGTGATCTCCGAGCAGACG |
| CmCHS2 | F:GACGAGACAAGTGTTGAGCGAGTA;R:TCCAGGTCCGAATCCAAAGAGC | |
| CmCHS3 | F:AGGAGCGATAGACGGGCACTTA;R:GAACAGCGAGTTCCAATCTAACATC | |
| CmCHS4 | F:GGAGGTGCTGAAGGAGTATGGGAACA;R:TCCAGGGCCGAACCCAAATAGA | |
| CmActin | F:GCATCACACCTTCTACAA;R:CATTGTAGAAGGTGTGATG | |
| 制备重组蛋白 Preparation of recombinant protein | CmCHS1 | F:TGGCTGATATCGGATCCATGGTGAGTATCGATGAG; R:CGACGGAGCTCGAATTCTTAATTAGTAGCCGCGCT |
| CmCHS2 | F:TGGCTGATATCGGATCCATGGTGAGTGTCCATGAG; R:CGACGGAGCTCGAATTCTTAAATAGCTACGCTGTG | |
| CmCHS3 | F:TGGCTGATATCGGATCCATGGTGAGCGTGGAAGAG; R:CGACGGAGCTCGAATTCTTAATTAGTAGCCACACT | |
| CmCHS4 | F:TGGCTGATATCGGATCCATGGCCACAATTGAGGAG; R:CGACGGAGCTCGAATTCTCAATTGATCGCCACACT | |
| 亚细胞定位 Subcellular | GFP(35S:GFP) | F:CTGATTACGCTCATATGATGGTGAGCAAGGGCGAG; R:AGGATTCAATCTTAAGTTACTTGTACAGCTCGTCCATG |
| localization | (GFP)CmCHS1 | F:GACGAGCTGTACAAGCATATGGTGAGTATCGAT; R:CAACAGGATTCAATCTTAAGTTAATTAGTAGCCGC |
| (GFP)CmCHS2 | F:GACGAGCTGTACAAGCATATGGTGAGTGTCCAT; R:CAACAGGATTCAATCTTAAGTTAAATAGCTACGCT | |
| (GFP)CmCHS3 | F:GACGAGCTGTACAAGCATATGGTGAGCGTGGAA; R:CAACAGGATTCAATCTTAAGTTAATTAGTAGCCAC | |
| (GFP)CmCHS4 | F:GACGAGCTGTACAAGCATATGGCCACAATTGAG; R:CAACAGGATTCAATCTTAAGTCAATTGATCGCCAC |

图1 君子兰(Clivia miniata)与其他物种CHS序列比对 AtCHS:拟南芥,AAA32771.1;FhCHS:香雪兰,JF732897;GbCHS:银杏,AAT68477.1;ZmCHS:玉米,CAA42763.1;MsCHS:紫花苜蓿,P30074.1;VvCHS:葡萄,BAA31259.1;VvSTS:葡萄,ABV82966.1。黄色框表示CHS中3个守恒催化残基;绿框中氨基酸决定CHS底物特异性;红色框中氨基酸为丙二酰辅酶A的结合区域;黑色矩形框为CHS的高度保守结构域;蓝色圆圈表示与香豆素酰辅酶A结合过程中重要的残基以及CHS环化反应的特异性残基。
Fig. 1 Sequence alignment of CHSs from Clivia miniata and other species AtCHS:Arabidopsis thaliana,AAA32771.1;FhCHS:Freesia hybrida,JF732897;GbCHS:Ginkgo biloba,AAT68477.1;ZmCHS:Zea mays,CAA42763.1;MsCHS:Medicago sativa,P30074.1;VvCHS:Vitis vinifera,BAA31259.1;VvSTS:Vitis vinifera,ABV82966.1.The yellow box represents three conserved catalytic residues in CHS. The green frame amino acid determines the specificity of CHS substrate;The red frame amino acid is the binding region of malonyl coenzyme A. The black rectangular box indicates the highly conserved domains of CHS. The blue circles represent important residues in binding to coumarinyl coenzyme A and residues specific to the cyclic reaction of CHS.
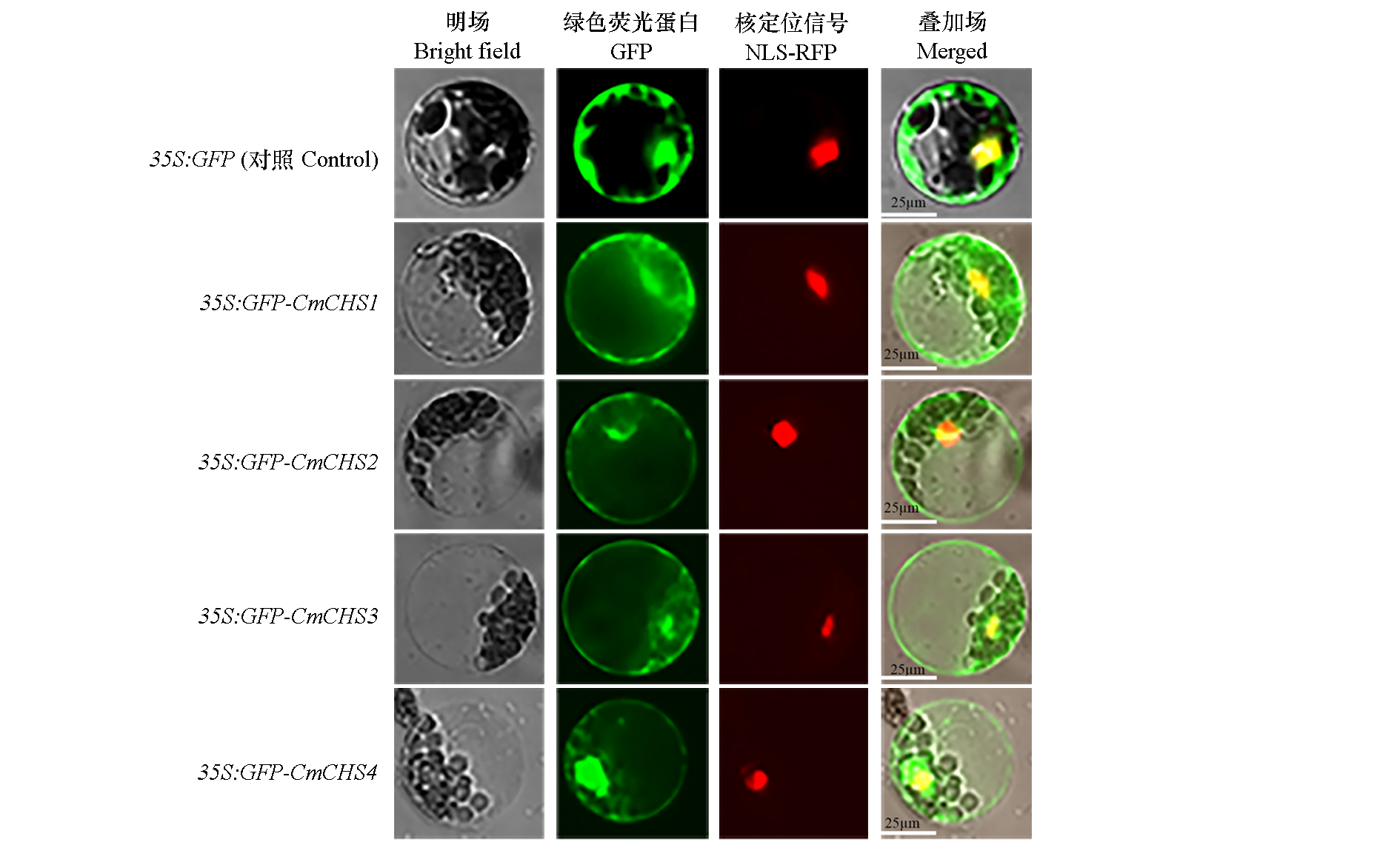
图3 CmCHS 亚细胞定位结果 将4个重组质粒瞬时转染到野生型拟南芥原生质体中,在室温黑暗中培养21 h。激光扫描共聚焦显微镜捕获荧光蛋白。
Fig. 3 Subcellular localization of CmCHSs Four recombinant plasmids were transiently transfected into wild-type Arabidopsis protoplasts,and incubated for 21 h at room temperature in the dark. Laser scanning confocal microscopy captured the fluorescent proteins.
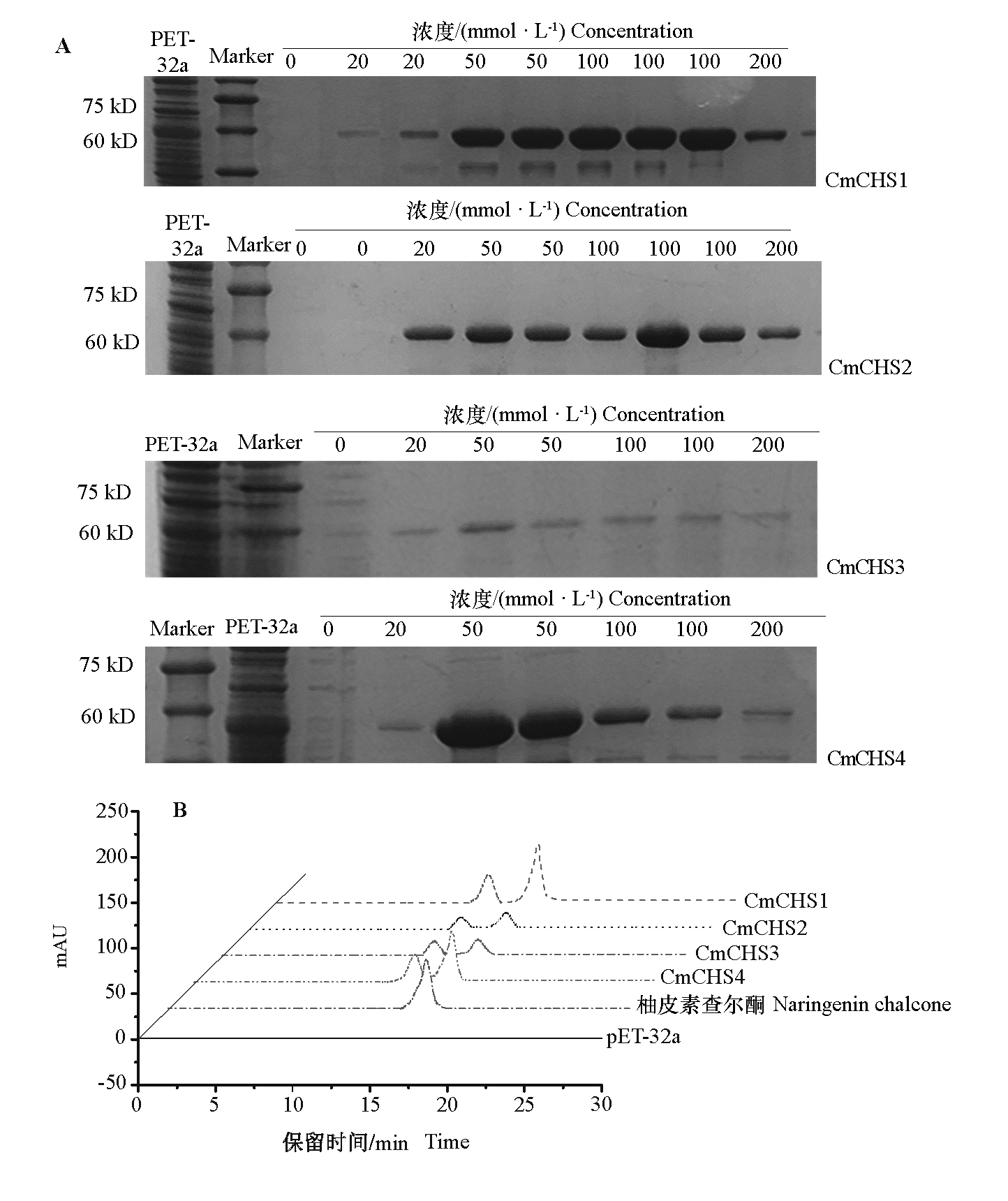
图5 CmCHS1、CmCHS2、CmCHS3和CmCHS4的体外酶学分析 A:提取重组蛋白SDS-PAGE;B:HPLC。
Fig. 5 In vitro enzymatic analysis of CmCHS1,CmCHS2,CmCHS3 and CmCHS4 A:The recombinant protein was extracted from sodium dodecyl sulfate polyacrylamide gel electrophoresis(SDS-PAGE);B:HPLC.
| [1] |
Abe I, Morita H. 2010. Structure and function of the chalcone synthase superfamily of plant type Ⅲ polyketide synthases. Natural Product Reports, 27 (6):809-838.
doi: 10.1039/b909988n URL |
| [2] |
Abe I, Takahashi Y, Noguchi H. 2002. Enzymatic formation of an unnatural C(6)-C(5) aromatic polyketide by plant type Ⅲ polyketide synthases. Organic Letters, 4 (21):3623-3626.
doi: 10.1021/ol0201409 URL |
| [3] |
Abe I, Utsumi Y, Oguro S, Noguchi H. 2004. The first plant type Ⅲ polyketide synthase that catalyzes formation of aromatic heptaketide. FEBS Letters, 562 (1-3):171-176.
doi: 10.1016/S0014-5793(04)00230-3 URL |
| [4] |
Agati G, Tattini M. 2010. Multiple functional roles of flavonoids in photoprotection. The New Phytologist, 186 (4):786-793.
doi: 10.1111/nph.2010.186.issue-4 URL |
| [5] | Aron P M, Kennedy J A. 2008. Flavan-3-ols:nature,occurrence and biological activity. Molecular Nutrition & Food Research, 52 (1):79-104. |
| [6] |
Austin M B, Noel J P. 2003. The chalcone synthase superfamily of type III polyketide synthases. Natural Product Reports, 20 (1):79-110.
doi: 10.1039/b100917f URL |
| [7] |
Deng X, Bashandy H, Ainasoja M, Kontturi J, Pietiäinen M, Laitinen R A, Albert V A, Valkonen J P, Elomaa P, Teeri T H. 2014. Functional diversification of duplicated chalcone synthase genes in anthocyanin biosynthesis of Gerbera hybrida. The New Phytologist, 201 (4):1469-1483.
doi: 10.1111/nph.2014.201.issue-4 URL |
| [8] |
Ferrer J L, Jez J M, Bowman M E, Dixon R A, Noel J P. 1999. Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nature Structural Biology, 6 (8):775-784.
pmid: 10426957 |
| [9] |
Forkmann G, Martens S. 2001. Metabolic engineering and applications of flavonoids. Current Opinion in Biotechnology, 12 (2):155-160.
pmid: 11287230 |
| [10] |
Franken P, Niesbach-Klösgen U, Weydemann U, Maréchal-Drouard L, Saedler H, Wienand U. 1991. The duplicated chalcone synthase genes C2and Whp(white pollen)of Zea mays are independently regulated;evidence for translational control of Whp expression by the anthocyanin intensifying gene in. The EMBO Journal, 10 (9):2605-2612.
doi: 10.1002/embj.1991.10.issue-9 URL |
| [11] |
Grotewold E. 2006. The genetics and biochemistry of floral pigments. Annual Review of Plant Biology, 57:761-780.
pmid: 16669781 |
| [12] |
Han Y, Zhao W, Wang Z, Zhu J, Liu Q. 2014. Molecular evolution and sequence divergence of plant chalcone synthase and chalcone synthase-like genes. Genetica, 142 (3):215-225.
doi: 10.1007/s10709-014-9768-3 URL |
| [13] |
Hermanns A S, Zhou X S, Xu Q, Tadmor Y, Li L. 2020. Carotenoid Pigment Accumulation in Horticultural Plants. Horticultural Plant Journal, 6 (6):343-360.
doi: 10.1016/j.hpj.2020.10.002 URL |
| [14] |
Jiang C, Schommer C K, Kim S Y, Suh D Y. 2006. Cloning and characterization of chalcone synthase from the moss,Physcomitrella patens. Phytochemistry, 67 (23):2531-2540.
doi: 10.1016/j.phytochem.2006.09.030 URL |
| [15] |
Joachim S. 1997. A family of plant-specific polyketide synthases:facts and predictions. Trends in Plant Science, 2 (10):373-378.
doi: 10.1016/S1360-1385(97)87121-X URL |
| [16] |
Kim S S, Grienenberger E, Lallemand B, Colpitts C C, Kim S Y, Souza C, Geoffroy P, Heintz D, Krahn D, Kaiser M, Kombrink E, Heitz T, Suh D Y, Legrand M, Douglas C J. 2010. Lap6/polyketide synthase a and lap5/polyketide synthase b encode hydroxyalkyl α-pyrone synthases required for pollen development and sporopollenin biosynthesis in Arabidopsis thaliana. The Plant Cell, 22 (12):4045-4066.
doi: 10.1105/tpc.110.080028 URL |
| [17] |
Koes R, Verweij W, Quattrocchio F. 2005. Flavonoids:a colorful model for the regulation and evolution of biochemical pathways. Trends in Plant Science, 10 (5):236-242.
doi: 10.1016/j.tplants.2005.03.002 URL |
| [18] |
Koes R E, Spelt C E, van den Elzen P J, Mol J N. 1989. Cloning and molecular characterization of the chalcone synthase multigene family of Petunia hybrida. Gene, 81 (2):245-257.
pmid: 2806915 |
| [19] |
Kong J M, Chia L S, Goh N K, Chia T F, Brouillard R. 2003. Analysis and biological activities of anthocyanins. Phytochemistry, 64 (5):923-933.
doi: 10.1016/S0031-9422(03)00438-2 URL |
| [20] |
Li Y, Shan X, Gao R, Han T, Zhang J, Wang Y, Kimani S, Wang L, Gao X. 2020. MYB repressors and MBW activation complex collaborate to fine-tune flower coloration in Freesia hybrida. Communications Biology, 3 (1):396.
doi: 10.1038/s42003-020-01134-6 URL |
| [21] |
Li Y, Shan X, Zhou L, Gao R, Yang S, Wang S, Wang L, Gao X. 2019. The R2R3-MYB Factor FhMYB 5 from Freesia hybrida contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis. Frontiers in Plant Science, 9:1935.
doi: 10.3389/fpls.2018.01935 URL |
| [22] |
Liou G, Chiang Y C, Wang Y, Weng J K. 2018. Mechanistic basis for the evolution of chalcone synthase catalytic cysteine reactivity in land plants. The Journal of Biological Chemistry, 293 (48):18601-18612.
doi: 10.1074/jbc.RA118.005695 URL |
| [23] |
Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCт method. Methods, 25 (4):402-408.
pmid: 11846609 |
| [24] |
Luan Y, Fu X, Lu P, Grierson D, Xu C. 2020. Molecular mechanisms determining the differential accumulation of carotenoids in plant species and varieties. Critical Reviews in Plant Sciences, 39 (2):1-15.
doi: 10.1080/07352689.2020.1724433 URL |
| [25] |
Morita H, Takahashi Y, Noguchi H, Abe I. 2000. Enzymatic formation of unnatural aromatic polyketides by chalcone synthase. Biochemical and Biophysical Research Communications, 279 (1):190-195.
pmid: 11112437 |
| [26] |
Morita Y, Saito R, Ban Y, Tanikawa N, Kuchitsu K, Ando T, Yoshikawa M, Habu Y, Ozeki Y, Nakayama M. 2012. Tandemly arranged chalcone synthase A genes contribute to the spatially regulated expression of siRNA and the natural bicolor floral phenotype in Petunia hybrida. The Plant Journal:for Cell and Molecular Biology, 70 (5):739-749.
doi: 10.1111/j.1365-313X.2012.04908.x URL |
| [27] |
Nakatsuka A, Izumi Y, Yamagishi M. 2003. Spatial and temporal expression of chalcone synthase and dihydroflavonol 4-reductase genes in the Asiatic hybrid lily. Plant Science, 165 (4):759-767.
doi: 10.1016/S0168-9452(03)00254-1 URL |
| [28] |
Napoli C A, Fahy D, Wang H Y, Taylor L P. 1999. White anther:a petunia mutant that abolishes pollen flavonol accumulation,induces male sterility,and is complemented by a chalcone synthase transgene. Plant Physiology, 120 (2):615-622.
pmid: 10364414 |
| [29] | Petroni K, Tonelli C. 2011. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Science:an International Journal of Experimental Plant Biology, 181 (3):219-229. |
| [30] |
Saigo T, Wang T, Watanabe M, Tohge T. 2020. Diversity of anthocyanin and proanthocyanin biosynthesis in land plants. Current Opinion in Plant Biology, 55:93-99.
doi: 10.1016/j.pbi.2020.04.001 URL |
| [31] |
Saito K, Yonekura-Sakakibara K, Nakabayashi R, Higashi Y, Yamazaki M, Tohge T, Fernie A R. 2013. The flavonoid biosynthetic pathway in Arabidopsis:structural and genetic diversity. Plant Physiology and Biochemistry, 72:21-34.
doi: 10.1016/j.plaphy.2013.02.001 URL |
| [32] |
Shan X, Li Y, Yang S, Gao R, Zhou L, Bao T, Han T, Wang S, Gao X, Wang L. 2019. A functional homologue of Arabidopsis TTG1 from Freesia interacts with bHLH proteins to regulate anthocyanin and proanthocyanidin biosynthesis in both Freesia hybrida and Arabidopsis thaliana. Plant Physiology and Biochemistry, 141:60-72.
doi: 10.1016/j.plaphy.2019.05.015 URL |
| [33] |
Shan X, Li Y, Yang S, Yang Z, Qiu M, Gao R, Han T, Meng X, Xu Z, Wang L, Gao X. 2020. The spatio-temporal biosynthesis of floral flavonols is controlled by differential phylogenetic MYB regulators in Freesia hybrida. New Phytologist, 228 (6):1864-1879.
doi: 10.1111/nph.v228.6 URL |
| [34] |
Shirley B W, Kubasek W L, Storz G, Bruggemann E, Koornneef M, Ausubel F M, Goodman H M. 1995. Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. The Plant Journal:For Cell and Molecular Biology, 8 (5):659-671.
doi: 10.1046/j.1365-313X.1995.08050659.x URL |
| [35] |
Sun W, Meng X, Liang L, Jiang W, Huang Y, He J, Hu H, Almqvist J, Gao X, Wang L. 2015. Molecular and biochemical analysis of chalcone synthase from Freesia hybrid in flavonoid biosynthetic pathway. PLoS ONE, 10 (3):e0119054.
doi: 10.1371/journal.pone.0119054 URL |
| [36] |
Tanaka Y, Ohmiya A. 2008. Seeing is believing: engineering anthocyanin and carotenoid biosynthetic pathways. Current Opinion in Biotechnology, 19 (2):190-197.
doi: 10.1016/j.copbio.2008.02.015 URL |
| [37] |
Tanaka Y, Sasaki N, Ohmiya A. 2008. Biosynthesis of plant pigments:anthocyanins,betalains and carotenoids. The Plant Journal, 54 (4):733-749.
doi: 10.1111/j.1365-313X.2008.03447.x URL |
| [38] |
Timoneda A, Feng T, Sheehan H, Walker-Hale N, Pucker B, Lopez-Nieves S, Guo R, Brockington S. 2019. The evolution of betalain biosynthesis in Caryophyllales. New Phytologist, 224 (1):71-85.
doi: 10.1111/nph.15980 pmid: 31172524 |
| [39] |
Václav T, Milena M, Tomáš V, Bořivoj K, Pavel H, Ladislav H. 2014. Chalcone synthase expression and pigments deposition in wheat with purple and blue colored caryopsis. Journal of Cereal Science, 59 (10):48-55.
doi: 10.1016/j.jcs.2013.10.008 URL |
| [40] |
Wang X, Wang X, Hu Q, Dai X, Tian H, Zheng K, Wang X, Mao T, Chen J G, Wang S. 2015. Characterization of an activation-tagged mutant uncovers a role of GLABRA 2 in anthocyanin biosynthesis in Arabidopsis. The Plant Journal, 83 (2):300-311.
doi: 10.1111/tpj.12887 URL |
| [41] |
Wang Z, Yu Q, Shen W, El Mohtar C A, Zhao X, Gmitter F G Jr. 2018. Functional study of CHS gene family members in citrus revealed a novel CHS gene affecting the production of flavonoids. BMC Plant Biology, 18 (1):189.
doi: 10.1186/s12870-018-1418-y URL |
| [42] |
Weng J K, Chapple C. 2010. The origin and evolution of lignin biosynthesis. The New Phytologist, 187 (2):273-285.
doi: 10.1111/nph.2010.187.issue-2 URL |
| [43] |
Winkel-Shirley B. 2001. Flavonoid biosynthesis. a colorful model for genetics,biochemistry,cell biology,and biotechnology. Plant Physiology, 126 (2):485-493.
pmid: 11402179 |
| [44] |
Yamazaki Y, Suh D Y, Sitthithaworn W, Ishiguro K, Kobayashi Y, Shibuya M, Ebizuka Y, Sankawa U. 2001. Diverse chalcone synthase superfamily enzymes from the most primitive vascular plant,Psilotum nudum. Planta, 214 (1):75-84.
pmid: 11762173 |
| [45] |
Yonekura-Sakakibara K, Higashi Y, Nakabayashi R. 2019. The origin and evolution of plant flavonoid metabolism. Frontiers in Plant Science, 10:943.
doi: 10.3389/fpls.2019.00943 pmid: 31428108 |
| [46] | Zhang Bi-xian, Zhu Yan-ming, Lai Yong-cai, Hu Xiao-mei, Li Wei, Li Wan, Bi Ying-dong, Xiao Jia-lei, Zhang Li-li. 2012. Research progress of plant chalcone synthase(CHS)and its gene. Journal of Anhui Agricultural Sciences, 40 (20):10376-10379.(in Chinese) |
| 张必弦, 朱延明, 来永才, 胡小梅, 李炜, 李琬, 毕影东, 肖佳雷, 张俐俐. 2012. 植物查尔酮合酶(CHS)及其基因的研究进展. 安徽农业科学, 40 (20):10376-10379. | |
| [47] | Zheng Yi, Zhang Hui, Wang Qinmei, Gao Yue, Zhang Zhihong, Sun Yuxin. 2020. Complete chloroplast genome sequence of Clivia miniata and its characteristics. Acta Horticulturae Sinica, 47 (12):2439-2450.(in Chinese) |
| 郑祎, 张卉, 王钦美, 高悦, 张志宏, 孙玉新. 2020. 大花君子兰叶绿体基因组及其特征. 园艺学报, 47 (12):2439-2450. | |
| [48] | Zhou L, Zheng K, Wang X, Tian H, Wang X, Wang S. 2014. Control of trichome formation in Arabidopsis by poplar single-repeat R3 MYB transcription factors. Frontiers in Plant Science, 5:262. |
| [49] |
Zhu K J, Wu Q J, Huang Y, Ye J L, Xu Q, Deng X X. 2020. Genome-wide characterization of cis-acting elements in the promoters of key carotenoid pathway genes from the main species of genus citrus. Horticultural Plant Journal, 6 (6):385-395.
doi: 10.1016/j.hpj.2020.10.003 URL |
| [1] | 叶子茂, 申晚霞, 刘梦雨, 王 彤, 张晓楠, 余 歆, 刘小丰, 赵晓春, . R2R3-MYB转录因子CitMYB21对柑橘类黄酮生物合成的影响[J]. 园艺学报, 2023, 50(2): 250-264. |
| [2] | 葛诗蓓, 张学宁, 韩文炎, 李青云, 李鑫. 植物类黄酮的生物合成及其抗逆作用机制研究进展[J]. 园艺学报, 2023, 50(1): 209-224. |
| [3] | 邹 雪, 丁 凡, 刘丽芳, 余韩开宗, 陈年伟, 饶莉萍. 紫色马铃薯新品种‘绵紫芋1号’[J]. 园艺学报, 2022, 49(S1): 93-94. |
| [4] | 黄玲, 胡先梅, 梁泽慧, 王艳平, 产祝龙, 向林. 郁金香花青素合成酶基因TgANS的克隆与功能鉴定[J]. 园艺学报, 2022, 49(9): 1935-1944. |
| [5] | 李茂福, 杨媛, 王华, 范又维, 孙佩, 金万梅. 月季中R2R3-MYB基因RhMYB113c调控花青素苷合成[J]. 园艺学报, 2022, 49(9): 1957-1966. |
| [6] | 许海峰, 王中堂, 陈新, 刘志国, 王利虎, 刘平, 刘孟军, 张琼. 冬枣果皮着色相关类黄酮靶向代谢组学及潜在MYB转录因子分析[J]. 园艺学报, 2022, 49(8): 1761-1771. |
| [7] | 钱婕妤, 蒋玲莉, 郑钢, 陈佳红, 赖吴浩, 许梦晗, 付建新, 张超. 百日草花青素苷合成相关MYB转录因子筛选及ZeMYB9功能研究[J]. 园艺学报, 2022, 49(7): 1505-1518. |
| [8] | 张鲁刚, 卢倩倩, 何琼, 薛一花, 马晓敏, 马帅, 聂姗姗, 杨文静. 紫橙色大白菜新种质的创制[J]. 园艺学报, 2022, 49(7): 1582-1588. |
| [9] | 陈道宗, 刘镒, 沈文杰, 朱博, 谭晨. 白菜、甘蓝和甘蓝型油菜PAP1/2同源基因的鉴定及分析[J]. 园艺学报, 2022, 49(6): 1301-1312. |
| [10] | 苏艳丽, 高晓铭, 杨健, 王龙, 王苏珂, 张向展, 薛华柏. 梨果实发育过程中褐变度、酚类物质及相关酶的变化研究[J]. 园艺学报, 2022, 49(11): 2304-2312. |
| [11] | 孙嘉茂, 崔全石, 王语晴, 司雅静, 时瑀繁, 卜海东, 袁晖, 王爱德. 苹果采前喷施EBR与MeJA对采后品质的影响[J]. 园艺学报, 2022, 49(10): 2236-2248. |
| [12] | 杨丽媛, 王倩, 王许会, 徐通达, 马军. 草莓生长素合成关键酶FveTAA1保守氨基酸位点T111的生物学功能研究[J]. 园艺学报, 2021, 48(9): 1695-1705. |
| [13] | 朱自果, 张庆田, 韩真, 李勃, 李国栋, 李秀杰. 欧洲葡萄VvMYB6正向调控花青素合成[J]. 园艺学报, 2021, 48(3): 465-476. |
| [14] | 马璐琳, 段青, 崔光芬, 杜文文, 贾文杰, 王祥宁, 王继华, 陈发棣. 钝裂银莲花花色素合成相关基因qRT-PCR内参基因的筛选[J]. 园艺学报, 2021, 48(2): 377-388. |
| [15] | 张晓楠, 余歆, 叶子茂, 刘小丰, 朱延松, 杨胜男, 王旭, 刘梦雨, 赵晓春. 宽皮柑橘果实的剥皮性及与细胞壁多糖的关系[J]. 园艺学报, 2021, 48(12): 2336-2348. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司