
园艺学报 ›› 2021, Vol. 48 ›› Issue (8): 1485-1503.doi: 10.16420/j.issn.0513-353x.2020-0595
张志强, 卢世雄, 马宗桓, 李彦彪, 高彩霞, 陈佰鸿, 毛娟( )
)
收稿日期:2021-01-22
修回日期:2021-04-06
出版日期:2021-08-25
发布日期:2021-09-06
通讯作者:
毛娟
E-mail:maojuan-81@163.com
基金资助:
ZHANG Zhiqiang, LU Shixiong, MA Zonghuan, LI Yanbiao, GAO Caixia, CHEN Baihong, MAO Juan( )
)
Received:2021-01-22
Revised:2021-04-06
Online:2021-08-25
Published:2021-09-06
Contact:
MAO Juan
E-mail:maojuan-81@163.com
摘要:
利用NCBI、GDR数据库对草莓LIM家族候选基因在草莓基因组水平进行生物信息学分析,鉴定获得4个亚族37个候选LIM基因,其中9个来自森林草莓(Fragaria vesca L.),28个来自凤梨草莓(F. ananassa Duch.)。草莓LIM家族候选基因主要分布在细胞核;其密码子偏好性较弱,偏向使用以A/T结尾的同义密码子。FvLIM和FaLIM启动子序列上游2 kb区域包含大量激素及非生物胁迫响应元件。10% PEG、200 mmol · L-1 NaCl、4 ℃低温胁迫下,随着处理时间延长LIM家族候选基因在凤梨草莓‘红颜’中比森林草莓中上调趋势更明显。4 ℃低温处理下,森林草莓中以FvLIM5(12 h)和FvLIM7(24 h)上调最为明显,而凤梨草莓‘红颜’中以FaLIM25(12 h)和FaLIM18(24 h)上调最为显著,总体对低温响应较明显。实时荧光定量PCR分析结果表明,草莓LIM家族候选基因在不同组织中的表达量存在一定差异,在根和花中的表达量偏高。综合以上分析结果,预测草莓LIM家族候选基因可能在响应非生物胁迫以及根和花的发育中发挥重要作用。
中图分类号:
张志强, 卢世雄, 马宗桓, 李彦彪, 高彩霞, 陈佰鸿, 毛娟. 草莓LIM家族候选基因的鉴定及在非生物胁迫下的表达[J]. 园艺学报, 2021, 48(8): 1485-1503.
ZHANG Zhiqiang, LU Shixiong, MA Zonghuan, LI Yanbiao, GAO Caixia, CHEN Baihong, MAO Juan. Bioinformatic Identification and Expression Analysis of Candidate Genes of LIM Protein Family in Strawberry Under Abiotic Stresses[J]. Acta Horticulturae Sinica, 2021, 48(8): 1485-1503.
| 基因 Gene | 上游引物(5′-3′) Forward primer | 下游引物(5′-3′) Reverse primer |
|---|---|---|
| FvLIM1 | ATATGTGGCTTGACGCAGAGACG | GAGGACGCTGAGGAGGAAGAGG |
| FvLIM2 | AAACACCACCACATCCAGCTCTTC | GGCTTGCTCGTTCGTTGATTGC |
| FvLIM3 | ATATGTGGCTTGACGCAGAGACG | GAGGACGCTGAGGAGGAAGAGG |
| FvLIM4 | GCACCACCACACCCAACTCATC | GCAACTGCTGTGGCTTTCTCATTG |
| FvLIM5 | TTGCCCAGTTGTTCAAGGAGAAGG | TGCTGCGTCTGGCTTTGGTTC |
| FvLIM6 | TGCAAGGCATGTGAGAAGACTGTG | AGTGGTGGCATCTGAAGCAAGC |
| FvLIM7 | GAATAGGGCACCCAGCAAACTCTC | TGAGCACACCTGAAGCATGACTTG |
| FvLIM8 | TTAGACCATGCAATCGCGCTCTC | GCTCGTTATCCTTCTCCTGCCATC |
| FvLIM9 | CTGGTATGTTTTCTGGCACGCAAG | TGGCTCTCCACTGTCACCTTCTC |
| FaLIM1 | ATATGTGGCTTGACGCAGAGACG | AAGAGGACGCTGAGGAGGAAGAC |
| FaLIM2 | GCCGAAGATTGGAGCAGATTACCG | ACGAGAATTGCCGTCACTTCACAG |
| FaLIM3 | ATATGTGGCTTGACGCAGAGACG | GAGGACGCTGAGGAGGAAGAGG |
| FaLIM4 | ATATGTGGCTTGACGCAGAGACG | AGGACGCTGAGAAGGAAGAGGAAG |
| FaLIM5 | GCACCACCACACCCAACTCATC | GCAACTGCTGTGGCTTTCTCATTG |
| FaLIM6 | GATAGGACTCCACGGTGTTGTAGC | TTCAAGGTAAAGGGGCTGGCATTC |
| FaLIM7 | GCTAGATGCCGAGCTTATGTCTGG | GCGTGCCCTTCTTTGATGAATTGG |
| FaLIM8 | AAACACCACCACATCCAGCTCTTC | GGCTTGCTCGTTCGTTGATTGC |
| FaLIM9 | TTGCCCAGTTGTTCAAGGAGAAGG | TGCTGCGTCTGGCTTTGGTTG |
| FaLIM10 | TTGCCCAGTTGTTCAAGGAGAAGG | TGCTGCGTCTGGCTTTGGTTC |
| FaLIM11 | CACCGCCTCTACTGTAAGCATCAC | TACTCCGCCACTGTGCTCTCAG |
| FaLIM12 | TTGCCCAGTTGTTCAAGGAGAAGG | TGCTGCGTCTGGCTTTGGTTC |
| FaLIM13 | TGCAAGGCATGTGAGAAGACTGTG | AGTGGTGGCATCTGAAGCAAGC |
| FaLIM14 | GAATAGGGCACCCAGCAAACTCTC | TGAGCACACCTGAAGCATGACTTG |
| FaLIM15 | AGTGCTTCTGTTGTCGCTCTTGTC | GGATGGGCCAGCTCCTTGAAAC |
| FaLIM16 | AGGAGAAGGGGAGCTACAACCATC | TGGAATGCTGGCAACTGCTACTG |
| FaLIM17 | GAATAGGGCACCCAGCAAACTCTC | TGAGCACACCTGAAGCATGACTTG |
| FaLIM18 | GAATAGGGCACCCAGCAAACTCTC | TGAGCACACCTGAAGCATGACTTG |
| FaLIM19 | TTAGACCATGCAATCGCGCTCTC | GCTCGTTATCCTTCTCCTGCCATC |
| FaLIM20 | CGACTAGGTCACCCAGCAAAGC | CGACTTGTGATATGCCTGGCTCTC |
| FaLIM21 | TTAGACCATGCAATCGCGCTCTC | GCTCGTTATCCTTCTCCTGCCATC |
| FaLIM22 | TCAAGGAGAAGGGGAGCTACAACC | TTAGGCTTCTGGAATGCTGGCAAC |
| FaLIM23 | TCAAGGAGAAGGGGAGCTACAACC | TTAGGCTTCTGGAATGCTGGCAAC |
| FaLIM24 | AGTGCTTCTGTTGTCGCTCTTGTC | GGATGGGCCAGCTCCTTGAAAC |
| FaLIM25 | AAGGTCCATTTCGCTCAGCTCTTC | CTTCTGGTTGTGGCTCTGCTTCTG |
| FaLIM26 | CTGGTATGTTTTCTGGCACGCAAG | TGGCTCTCCACTGTCACCTTCTC |
| FaLIM27 | AGTGCTTCTGTTGTCGCTCTTGTG | GGATGGGCCAGCTCCTTGAAAC |
| FaLIM28 | GAATAGGGCACCCAGCAAACTCTC | TGAGCACACCTGAAGCATGACTTG |
| GAPDH | CATTCATCACCACCGACTACA | GAAGGGTCTTCTCATCCTTGAC |
表1 草莓LIM家族候选基因实时荧光定量引物
Table 1 Real-time PCR primers for strawberry LIM family candidate genes
| 基因 Gene | 上游引物(5′-3′) Forward primer | 下游引物(5′-3′) Reverse primer |
|---|---|---|
| FvLIM1 | ATATGTGGCTTGACGCAGAGACG | GAGGACGCTGAGGAGGAAGAGG |
| FvLIM2 | AAACACCACCACATCCAGCTCTTC | GGCTTGCTCGTTCGTTGATTGC |
| FvLIM3 | ATATGTGGCTTGACGCAGAGACG | GAGGACGCTGAGGAGGAAGAGG |
| FvLIM4 | GCACCACCACACCCAACTCATC | GCAACTGCTGTGGCTTTCTCATTG |
| FvLIM5 | TTGCCCAGTTGTTCAAGGAGAAGG | TGCTGCGTCTGGCTTTGGTTC |
| FvLIM6 | TGCAAGGCATGTGAGAAGACTGTG | AGTGGTGGCATCTGAAGCAAGC |
| FvLIM7 | GAATAGGGCACCCAGCAAACTCTC | TGAGCACACCTGAAGCATGACTTG |
| FvLIM8 | TTAGACCATGCAATCGCGCTCTC | GCTCGTTATCCTTCTCCTGCCATC |
| FvLIM9 | CTGGTATGTTTTCTGGCACGCAAG | TGGCTCTCCACTGTCACCTTCTC |
| FaLIM1 | ATATGTGGCTTGACGCAGAGACG | AAGAGGACGCTGAGGAGGAAGAC |
| FaLIM2 | GCCGAAGATTGGAGCAGATTACCG | ACGAGAATTGCCGTCACTTCACAG |
| FaLIM3 | ATATGTGGCTTGACGCAGAGACG | GAGGACGCTGAGGAGGAAGAGG |
| FaLIM4 | ATATGTGGCTTGACGCAGAGACG | AGGACGCTGAGAAGGAAGAGGAAG |
| FaLIM5 | GCACCACCACACCCAACTCATC | GCAACTGCTGTGGCTTTCTCATTG |
| FaLIM6 | GATAGGACTCCACGGTGTTGTAGC | TTCAAGGTAAAGGGGCTGGCATTC |
| FaLIM7 | GCTAGATGCCGAGCTTATGTCTGG | GCGTGCCCTTCTTTGATGAATTGG |
| FaLIM8 | AAACACCACCACATCCAGCTCTTC | GGCTTGCTCGTTCGTTGATTGC |
| FaLIM9 | TTGCCCAGTTGTTCAAGGAGAAGG | TGCTGCGTCTGGCTTTGGTTG |
| FaLIM10 | TTGCCCAGTTGTTCAAGGAGAAGG | TGCTGCGTCTGGCTTTGGTTC |
| FaLIM11 | CACCGCCTCTACTGTAAGCATCAC | TACTCCGCCACTGTGCTCTCAG |
| FaLIM12 | TTGCCCAGTTGTTCAAGGAGAAGG | TGCTGCGTCTGGCTTTGGTTC |
| FaLIM13 | TGCAAGGCATGTGAGAAGACTGTG | AGTGGTGGCATCTGAAGCAAGC |
| FaLIM14 | GAATAGGGCACCCAGCAAACTCTC | TGAGCACACCTGAAGCATGACTTG |
| FaLIM15 | AGTGCTTCTGTTGTCGCTCTTGTC | GGATGGGCCAGCTCCTTGAAAC |
| FaLIM16 | AGGAGAAGGGGAGCTACAACCATC | TGGAATGCTGGCAACTGCTACTG |
| FaLIM17 | GAATAGGGCACCCAGCAAACTCTC | TGAGCACACCTGAAGCATGACTTG |
| FaLIM18 | GAATAGGGCACCCAGCAAACTCTC | TGAGCACACCTGAAGCATGACTTG |
| FaLIM19 | TTAGACCATGCAATCGCGCTCTC | GCTCGTTATCCTTCTCCTGCCATC |
| FaLIM20 | CGACTAGGTCACCCAGCAAAGC | CGACTTGTGATATGCCTGGCTCTC |
| FaLIM21 | TTAGACCATGCAATCGCGCTCTC | GCTCGTTATCCTTCTCCTGCCATC |
| FaLIM22 | TCAAGGAGAAGGGGAGCTACAACC | TTAGGCTTCTGGAATGCTGGCAAC |
| FaLIM23 | TCAAGGAGAAGGGGAGCTACAACC | TTAGGCTTCTGGAATGCTGGCAAC |
| FaLIM24 | AGTGCTTCTGTTGTCGCTCTTGTC | GGATGGGCCAGCTCCTTGAAAC |
| FaLIM25 | AAGGTCCATTTCGCTCAGCTCTTC | CTTCTGGTTGTGGCTCTGCTTCTG |
| FaLIM26 | CTGGTATGTTTTCTGGCACGCAAG | TGGCTCTCCACTGTCACCTTCTC |
| FaLIM27 | AGTGCTTCTGTTGTCGCTCTTGTG | GGATGGGCCAGCTCCTTGAAAC |
| FaLIM28 | GAATAGGGCACCCAGCAAACTCTC | TGAGCACACCTGAAGCATGACTTG |
| GAPDH | CATTCATCACCACCGACTACA | GAAGGGTCTTCTCATCCTTGAC |
| 名称 Name | 染色体定位 Chromosome location | 氨基酸数 Amino acids | 分子量 Molecular weight | 等电点 pI | 不稳定指数 Instability index | 亲水性 Hydrophilic | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|
| FvLIM1 | Fvb1:7945940..7952121 | 538 | 60 510.36 | 5.20 | 53.78 | -0.718 | 细胞核 Nucleus |
| FvLIM2 | Fvb2:2982770..2998244 | 197 | 22 232.22 | 8.89 | 42.90 | -0.703 | 细胞核 Nucleus |
| FvLIM3 | Fvb2:5932861..5935114 | 527 | 59 323.18 | 5.33 | 52.19 | -0.698 | 细胞核 Nucleus |
| FvLIM4 | Fvb2:24843809..24848493 | 191 | 21 205.29 | 9.05 | 29.01 | -0.539 | 细胞核 Nucleus |
| FvLIM5 | Fvb5:1235177..1237221 | 212 | 23 391.44 | 7.52 | 45.56 | -0.62 | 细胞核 Nucleus |
| FvLIM6 | Fvb5:9381785..9385122 | 188 | 21 211.15 | 8.86 | 28.77 | -0.586 | 细胞核 Nucleus |
| FvLIM7 | Fvb7:12341249..12343536 | 217 | 23 962.13 | 6.50 | 45.65 | -0.567 | 细胞核 Nucleus |
| FvLIM8 | Fvb7:18342947..18346948 | 504 | 57 014.81 | 8.26 | 52.67 | -0.541 | 细胞核Nucleus |
| FvLIM9 | Fvb7:18517083..18519868 | 200 | 21 721.91 | 9.08 | 37.48 | -0.362 | 细胞核 Nucleus |
| FaLIM1 | Fvb1-1:20431547..20437334 | 534 | 59 915.92 | 5.22 | 52.12 | -0.668 | 细胞核 Nucleus |
| FaLIM2 | Fvb1-2:9164763..9171264 | 533 | 60 168.24 | 5.39 | 51.21 | -0.737 | 细胞核 Nucleus |
| FaLIM3 | Fvb1-3:8145568..8151824 | 539 | 60 522.40 | 5.20 | 54.39 | -0.719 | 细胞核 Nucleus |
| FaLIM4 | Fvb1-4:6945308..6951056 | 538 | 60 571.45 | 5.17 | 53.06 | -0.712 | 细胞核 Nucleus |
| FaLIM5 | Fvb2-2:728488..730564 | 191 | 21 205.29 | 9.05 | 29.01 | -0.539 | 细胞核 Nucleus |
| FaLIM6 | Fvb2-2:18290991..18293017 | 231 | 27 087.33 | 7.80 | 45.08 | -0.512 | 细胞核 Nucleus |
| FaLIM7 | Fvb2-3:2848239..2854332 | 491 | 55 136.60 | 5.75 | 44.43 | -0.671 | 细胞核 Nucleus |
| FaLIM8 | Fvb2-4:1747334..1750087 | 176 | 19 899.54 | 8.83 | 44.58 | -0.618 | 细胞核 Nucleus |
| FaLIM9 | Fvb5-1:1118664..1120389 | 213 | 23 551.67 | 7.92 | 45.67 | -0.623 | 细胞核 Nucleus |
| FaLIM10 | Fvb5-2:1037693..1039462 | 216 | 23 835.04 | 8.18 | 44.28 | -0.569 | 细胞核 Nucleus |
| FaLIM11 | Fvb5-3:18813760..18816438 | 188 | 21 172.07 | 8.86 | 31.15 | -0.599 | 细胞核 Nucleus |
| FaLIM12 | Fvb5-4:1027068..1028886 | 211 | 23 297.40 | 7.52 | 43.54 | -0.607 | 细胞核 Nucleus |
| FaLIM13 | Fvb5-4:7252377..7255038 | 188 | 21 199.10 | 8.86 | 29.68 | -0.613 | 细胞核 Nucleus |
| FaLIM14 | Fvb7-1:16915680..16917768 | 217 | 23 964.10 | 6.50 | 45.95 | -0.579 | 细胞核 Nucleus |
| FaLIM15 | Fvb7-1:25526059..25530050 | 506 | 57 362.23 | 8.54 | 52.15 | -0.549 | 细胞核Nucleus |
| FaLIM16 | Fvb7-1:25665993..25670535 | 200 | 21 818.02 | 9.08 | 37.58 | -0.360 | 细胞核 Nucleus |
| FaLIM17 | Fvb7-2:11429297..11431530 | 217 | 23 962.13 | 6.50 | 45.65 | -0.567 | 细胞核 Nucleus |
| FaLIM18 | Fvb7-2:17869656..17871925 | 217 | 23 949.13 | 6.50 | 45.65 | -0.554 | 细胞核 Nucleus |
| FaLIM19 | Fvb7-2:23292270..23296225 | 493 | 55 877.59 | 8.37 | 53.86 | -0.540 | 细胞核 Nucleus |
| FaLIM20 | Fvb7-2:23449921..23452679 | 200 | 21 691.82 | 9.08 | 37.63 | -0.376 | 细胞核 Nucleus |
| FaLIM21 | Fvb7-2:23655808..23659719 | 430 | 49 337.54 | 8.63 | 48.07 | -0.525 | 细胞核Nucleus |
| FaLIM22 | Fvb7-2:23849991..23852934 | 200 | 21 721.91 | 9.08 | 37.48 | -0.362 | 细胞核 Nucleus |
| FaLIM23 | Fvb7-3:5169640..5172605 | 200 | 21 704.86 | 9.01 | 38.00 | -0.353 | 细胞核 Nucleus |
| FaLIM24 | Fvb7-3:6773453..6777457 | 506 | 57 325.21 | 8.45 | 52.87 | -0.541 | 细胞核Nucleus |
| FaLIM25 | Fvb7-3:12069028..12071004 | 215 | 23 793.94 | 6.50 | 44.19 | -0.573 | 细胞核 Nucleus |
| FaLIM26 | Fvb7-4:5056752..5059611 | 200 | 21 721.91 | 9.08 | 37.48 | -0.362 | 细胞核 Nucleus |
| FaLIM27 | Fvb7-4:5292185..5296170 | 506 | 57 277.11 | 8.45 | 51.10 | -0.545 | 细胞核Nucleus |
| FaLIM28 | Fvb7-4:11548941..11551124 | 214 | 23 743.92 | 6.54 | 42.18 | -0.534 | 细胞核 Nucleus |
表2 草莓LIM家族候选基因理化性质
Table 2 Physical and chemical properties for strawberry LIM family candidate genes
| 名称 Name | 染色体定位 Chromosome location | 氨基酸数 Amino acids | 分子量 Molecular weight | 等电点 pI | 不稳定指数 Instability index | 亲水性 Hydrophilic | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|
| FvLIM1 | Fvb1:7945940..7952121 | 538 | 60 510.36 | 5.20 | 53.78 | -0.718 | 细胞核 Nucleus |
| FvLIM2 | Fvb2:2982770..2998244 | 197 | 22 232.22 | 8.89 | 42.90 | -0.703 | 细胞核 Nucleus |
| FvLIM3 | Fvb2:5932861..5935114 | 527 | 59 323.18 | 5.33 | 52.19 | -0.698 | 细胞核 Nucleus |
| FvLIM4 | Fvb2:24843809..24848493 | 191 | 21 205.29 | 9.05 | 29.01 | -0.539 | 细胞核 Nucleus |
| FvLIM5 | Fvb5:1235177..1237221 | 212 | 23 391.44 | 7.52 | 45.56 | -0.62 | 细胞核 Nucleus |
| FvLIM6 | Fvb5:9381785..9385122 | 188 | 21 211.15 | 8.86 | 28.77 | -0.586 | 细胞核 Nucleus |
| FvLIM7 | Fvb7:12341249..12343536 | 217 | 23 962.13 | 6.50 | 45.65 | -0.567 | 细胞核 Nucleus |
| FvLIM8 | Fvb7:18342947..18346948 | 504 | 57 014.81 | 8.26 | 52.67 | -0.541 | 细胞核Nucleus |
| FvLIM9 | Fvb7:18517083..18519868 | 200 | 21 721.91 | 9.08 | 37.48 | -0.362 | 细胞核 Nucleus |
| FaLIM1 | Fvb1-1:20431547..20437334 | 534 | 59 915.92 | 5.22 | 52.12 | -0.668 | 细胞核 Nucleus |
| FaLIM2 | Fvb1-2:9164763..9171264 | 533 | 60 168.24 | 5.39 | 51.21 | -0.737 | 细胞核 Nucleus |
| FaLIM3 | Fvb1-3:8145568..8151824 | 539 | 60 522.40 | 5.20 | 54.39 | -0.719 | 细胞核 Nucleus |
| FaLIM4 | Fvb1-4:6945308..6951056 | 538 | 60 571.45 | 5.17 | 53.06 | -0.712 | 细胞核 Nucleus |
| FaLIM5 | Fvb2-2:728488..730564 | 191 | 21 205.29 | 9.05 | 29.01 | -0.539 | 细胞核 Nucleus |
| FaLIM6 | Fvb2-2:18290991..18293017 | 231 | 27 087.33 | 7.80 | 45.08 | -0.512 | 细胞核 Nucleus |
| FaLIM7 | Fvb2-3:2848239..2854332 | 491 | 55 136.60 | 5.75 | 44.43 | -0.671 | 细胞核 Nucleus |
| FaLIM8 | Fvb2-4:1747334..1750087 | 176 | 19 899.54 | 8.83 | 44.58 | -0.618 | 细胞核 Nucleus |
| FaLIM9 | Fvb5-1:1118664..1120389 | 213 | 23 551.67 | 7.92 | 45.67 | -0.623 | 细胞核 Nucleus |
| FaLIM10 | Fvb5-2:1037693..1039462 | 216 | 23 835.04 | 8.18 | 44.28 | -0.569 | 细胞核 Nucleus |
| FaLIM11 | Fvb5-3:18813760..18816438 | 188 | 21 172.07 | 8.86 | 31.15 | -0.599 | 细胞核 Nucleus |
| FaLIM12 | Fvb5-4:1027068..1028886 | 211 | 23 297.40 | 7.52 | 43.54 | -0.607 | 细胞核 Nucleus |
| FaLIM13 | Fvb5-4:7252377..7255038 | 188 | 21 199.10 | 8.86 | 29.68 | -0.613 | 细胞核 Nucleus |
| FaLIM14 | Fvb7-1:16915680..16917768 | 217 | 23 964.10 | 6.50 | 45.95 | -0.579 | 细胞核 Nucleus |
| FaLIM15 | Fvb7-1:25526059..25530050 | 506 | 57 362.23 | 8.54 | 52.15 | -0.549 | 细胞核Nucleus |
| FaLIM16 | Fvb7-1:25665993..25670535 | 200 | 21 818.02 | 9.08 | 37.58 | -0.360 | 细胞核 Nucleus |
| FaLIM17 | Fvb7-2:11429297..11431530 | 217 | 23 962.13 | 6.50 | 45.65 | -0.567 | 细胞核 Nucleus |
| FaLIM18 | Fvb7-2:17869656..17871925 | 217 | 23 949.13 | 6.50 | 45.65 | -0.554 | 细胞核 Nucleus |
| FaLIM19 | Fvb7-2:23292270..23296225 | 493 | 55 877.59 | 8.37 | 53.86 | -0.540 | 细胞核 Nucleus |
| FaLIM20 | Fvb7-2:23449921..23452679 | 200 | 21 691.82 | 9.08 | 37.63 | -0.376 | 细胞核 Nucleus |
| FaLIM21 | Fvb7-2:23655808..23659719 | 430 | 49 337.54 | 8.63 | 48.07 | -0.525 | 细胞核Nucleus |
| FaLIM22 | Fvb7-2:23849991..23852934 | 200 | 21 721.91 | 9.08 | 37.48 | -0.362 | 细胞核 Nucleus |
| FaLIM23 | Fvb7-3:5169640..5172605 | 200 | 21 704.86 | 9.01 | 38.00 | -0.353 | 细胞核 Nucleus |
| FaLIM24 | Fvb7-3:6773453..6777457 | 506 | 57 325.21 | 8.45 | 52.87 | -0.541 | 细胞核Nucleus |
| FaLIM25 | Fvb7-3:12069028..12071004 | 215 | 23 793.94 | 6.50 | 44.19 | -0.573 | 细胞核 Nucleus |
| FaLIM26 | Fvb7-4:5056752..5059611 | 200 | 21 721.91 | 9.08 | 37.48 | -0.362 | 细胞核 Nucleus |
| FaLIM27 | Fvb7-4:5292185..5296170 | 506 | 57 277.11 | 8.45 | 51.10 | -0.545 | 细胞核Nucleus |
| FaLIM28 | Fvb7-4:11548941..11551124 | 214 | 23 743.92 | 6.54 | 42.18 | -0.534 | 细胞核 Nucleus |
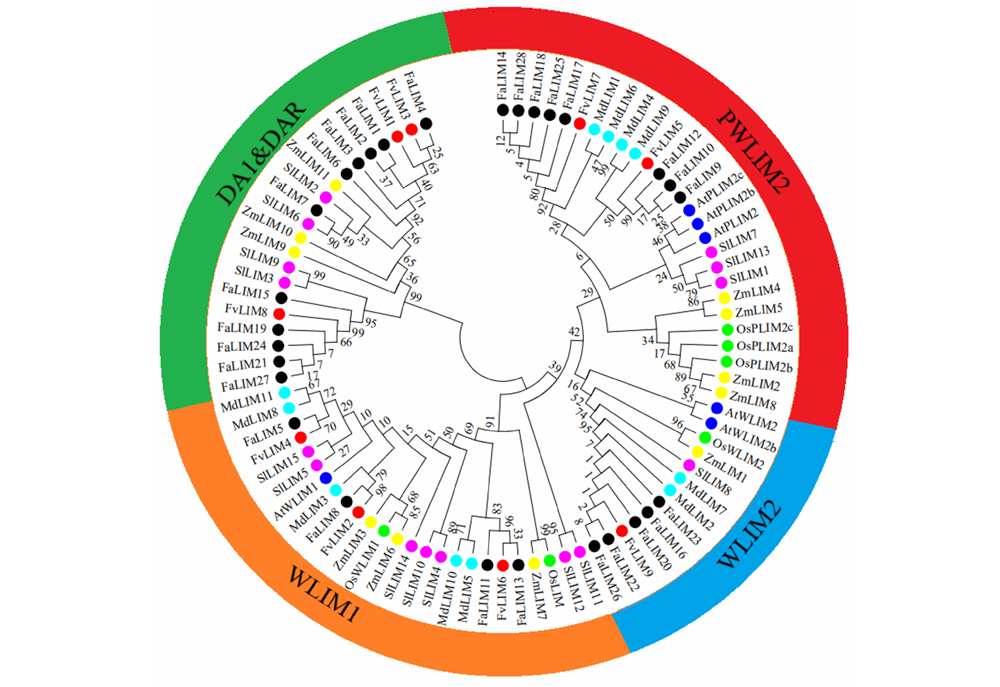
图1 草莓LIM家族候选基因进化树 Fa:凤梨草莓;Fv:森林草莓;Md:苹果;At:拟南芥;Sl:番茄;Zm:玉米;Os:水稻。
Fig. 1 The phylogenetic tree for strawberry LIM family candidate genes Fa:Fragaria ananassa;Fv:Fragaria vesca;Md:Malus × domestica;At:Arabidopsis thaliana;Sl:Solanum lycopersicum;Zm:Zea mays;Os:Oryza sativa.
| 基序编号 Conserved motif | 氨基酸数 Number of amino acids | 氨基酸保守序列 Amino acid conserved sequence |
|---|---|---|
| motif1 | 50 | GAVYHKSCFKCHHCKGPJKLSNYSSSEGVLYCKPHFEZLFKETGNVCKSF |
| motif2 | 50 | VYPLEKVTVEGESYHKSCFRCTHGGCPJTPSNYAALEGILYCKHHFAQLF |
| motif3 | 50 | AIMDTHECQPLYLEIQEFYEGLNMKVEQQVPMLLVERQALNEAMVGEKNG |
| motif4 | 50 | IDMRTQPQKLTRRCEVTAILVLYGLPRLLTGSILAHEMMHAWLRLKGYPN |
| motif5 | 50 | IPTNSAGLIEYRAHPFWLQKYCPSHERDRTPRCCSCERMEPKDTKYJLLD |
| motif6 | 21 | FPGTRQKCKACEKTVYLVDKL |
| motif7 | 50 | EKKLGEFFKHQIESDSSPAYGEGFRAGNQAVLKYGLRRTLDHIRITGSFP |
| motif8 | 21 | PSKLSSMFSGTQDKCATCKKT |
表3 草莓LIM家族候选基因氨基酸保守序列
Table 3 Conserved motifs for strawberry LIM family candidate genes
| 基序编号 Conserved motif | 氨基酸数 Number of amino acids | 氨基酸保守序列 Amino acid conserved sequence |
|---|---|---|
| motif1 | 50 | GAVYHKSCFKCHHCKGPJKLSNYSSSEGVLYCKPHFEZLFKETGNVCKSF |
| motif2 | 50 | VYPLEKVTVEGESYHKSCFRCTHGGCPJTPSNYAALEGILYCKHHFAQLF |
| motif3 | 50 | AIMDTHECQPLYLEIQEFYEGLNMKVEQQVPMLLVERQALNEAMVGEKNG |
| motif4 | 50 | IDMRTQPQKLTRRCEVTAILVLYGLPRLLTGSILAHEMMHAWLRLKGYPN |
| motif5 | 50 | IPTNSAGLIEYRAHPFWLQKYCPSHERDRTPRCCSCERMEPKDTKYJLLD |
| motif6 | 21 | FPGTRQKCKACEKTVYLVDKL |
| motif7 | 50 | EKKLGEFFKHQIESDSSPAYGEGFRAGNQAVLKYGLRRTLDHIRITGSFP |
| motif8 | 21 | PSKLSSMFSGTQDKCATCKKT |
| 基因Gene | T3s | C3s | A3s | G3s | CAI | Fop | ENC | GC3s | GC |
|---|---|---|---|---|---|---|---|---|---|
| FvLIM1 | 0.48 | 0.18 | 0.41 | 0.23 | 0.211 | 0.37 | 51.41 | 0.31 | 0.43 |
| FvLIM3 | 0.48 | 0.18 | 0.41 | 0.23 | 0.210 | 0.37 | 51.69 | 0.31 | 0.43 |
| FvLIM8 | 0.48 | 0.20 | 0.37 | 0.22 | 0.192 | 0.36 | 51.25 | 0.32 | 0.44 |
| FaLIM4 | 0.48 | 0.18 | 0.41 | 0.23 | 0.212 | 0.37 | 51.64 | 0.31 | 0.43 |
| FaLIM2 | 0.48 | 0.18 | 0.41 | 0.24 | 0.214 | 0.37 | 51.73 | 0.32 | 0.43 |
| FaLIM3 | 0.47 | 0.19 | 0.41 | 0.23 | 0.212 | 0.37 | 51.89 | 0.31 | 0.44 |
| FaLIM1 | 0.48 | 0.18 | 0.41 | 0.23 | 0.209 | 0.36 | 51.19 | 0.31 | 0.44 |
| FaLIM6 | 0.49 | 0.20 | 0.42 | 0.24 | 0.201 | 0.31 | 56.42 | 0.32 | 0.42 |
| FaLIM7 | 0.43 | 0.22 | 0.40 | 0.25 | 0.169 | 0.34 | 54.45 | 0.35 | 0.44 |
| FaLIM15 | 0.48 | 0.20 | 0.37 | 0.22 | 0.195 | 0.37 | 51.76 | 0.32 | 0.44 |
| FaLIM19 | 0.48 | 0.20 | 0.37 | 0.22 | 0.191 | 0.36 | 50.99 | 0.32 | 0.44 |
| FaLIM27 | 0.48 | 0.20 | 0.37 | 0.22 | 0.193 | 0.36 | 51.75 | 0.32 | 0.44 |
| FaLIM24 | 0.47 | 0.21 | 0.37 | 0.23 | 0.192 | 0.37 | 52.14 | 0.33 | 0.45 |
| FvLIM21 | 0.48 | 0.20 | 0.36 | 0.24 | 0.181 | 0.35 | 50.30 | 0.33 | 0.44 |
| FvLIM7 | 0.38 | 0.34 | 0.38 | 0.24 | 0.266 | 0.42 | 43.11 | 0.43 | 0.46 |
| FaLIM17 | 0.38 | 0.34 | 0.38 | 0.23 | 0.268 | 0.43 | 43.14 | 0.43 | 0.46 |
| FaLIM18 | 0.38 | 0.34 | 0.37 | 0.24 | 0.270 | 0.43 | 42.82 | 0.43 | 0.46 |
| FaLIM14 | 0.42 | 0.33 | 0.34 | 0.25 | 0.262 | 0.42 | 43.15 | 0.43 | 0.46 |
| FaLIM28 | 0.41 | 0.33 | 0.34 | 0.25 | 0.260 | 0.43 | 44.11 | 0.44 | 0.46 |
| FaLIM25 | 0.41 | 0.32 | 0.37 | 0.24 | 0.273 | 0.44 | 41.34 | 0.42 | 0.46 |
| FvLIM5 | 0.33 | 0.38 | 0.35 | 0.28 | 0.271 | 0.45 | 52.09 | 0.49 | 0.48 |
| 基因Gene | T3s | C3s | A3s | G3s | CAI | Fop | ENC | GC3s | GC |
| FaLIM10 | 0.32 | 0.39 | 0.34 | 0.28 | 0.273 | 0.46 | 60.52 | 0.50 | 0.48 |
| FaLIM12 | 0.32 | 0.39 | 0.34 | 0.28 | 0.272 | 0.45 | 55.80 | 0.50 | 0.48 |
| FaLIM9 | 0.32 | 0.38 | 0.35 | 0.29 | 0.265 | 0.44 | 51.91 | 0.50 | 0.47 |
| FaLIM22 | 0.39 | 0.32 | 0.29 | 0.32 | 0.334 | 0.45 | 56.12 | 0.48 | 0.47 |
| FaLIM23 | 0.39 | 0.31 | 0.28 | 0.33 | 0.348 | 0.47 | 52.97 | 0.48 | 0.47 |
| FaLIM20 | 0.39 | 0.31 | 0.29 | 0.32 | 0.333 | 0.45 | 55.85 | 0.48 | 0.47 |
| FvLIM9 | 0.39 | 0.32 | 0.29 | 0.32 | 0.332 | 0.44 | 55.51 | 0.48 | 0.47 |
| FaLIM26 | 0.39 | 0.32 | 0.29 | 0.32 | 0.332 | 0.44 | 55.51 | 0.48 | 0.47 |
| FaLIM16 | 0.39 | 0.31 | 0.29 | 0.32 | 0.351 | 0.46 | 52.26 | 0.47 | 0.47 |
| FvLIM2 | 0.29 | 0.42 | 0.34 | 0.30 | 0.266 | 0.46 | 58.30 | 0.53 | 0.47 |
| FaLIM8 | 0.27 | 0.44 | 0.34 | 0.31 | 0.242 | 0.44 | 59.77 | 0.55 | 0.48 |
| FvLIM4 | 0.29 | 0.41 | 0.38 | 0.24 | 0.254 | 0.48 | 51.73 | 0.50 | 0.47 |
| FaLIM5 | 0.29 | 0.42 | 0.38 | 0.23 | 0.259 | 0.48 | 51.62 | 0.50 | 0.47 |
| FvLIM6 | 0.40 | 0.35 | 0.28 | 0.33 | 0.318 | 0.48 | 59.48 | 0.50 | 0.45 |
| FaLIM13 | 0.41 | 0.34 | 0.29 | 0.32 | 0.316 | 0.47 | 56.22 | 0.48 | 0.45 |
| FaLIM11 | 0.40 | 0.36 | 0.28 | 0.31 | 0.320 | 0.48 | 55.92 | 0.49 | 0.45 |
| 平均 Average | 0.41 | 0.29 | 0.35 | 0.26 | 0.256 | 0.42 | 52.10 | 0.42 | 0.45 |
表4 草莓LIM家族候选基因组主要参数
Table 4 Main parameters for strawberry LIM family candidate genes
| 基因Gene | T3s | C3s | A3s | G3s | CAI | Fop | ENC | GC3s | GC |
|---|---|---|---|---|---|---|---|---|---|
| FvLIM1 | 0.48 | 0.18 | 0.41 | 0.23 | 0.211 | 0.37 | 51.41 | 0.31 | 0.43 |
| FvLIM3 | 0.48 | 0.18 | 0.41 | 0.23 | 0.210 | 0.37 | 51.69 | 0.31 | 0.43 |
| FvLIM8 | 0.48 | 0.20 | 0.37 | 0.22 | 0.192 | 0.36 | 51.25 | 0.32 | 0.44 |
| FaLIM4 | 0.48 | 0.18 | 0.41 | 0.23 | 0.212 | 0.37 | 51.64 | 0.31 | 0.43 |
| FaLIM2 | 0.48 | 0.18 | 0.41 | 0.24 | 0.214 | 0.37 | 51.73 | 0.32 | 0.43 |
| FaLIM3 | 0.47 | 0.19 | 0.41 | 0.23 | 0.212 | 0.37 | 51.89 | 0.31 | 0.44 |
| FaLIM1 | 0.48 | 0.18 | 0.41 | 0.23 | 0.209 | 0.36 | 51.19 | 0.31 | 0.44 |
| FaLIM6 | 0.49 | 0.20 | 0.42 | 0.24 | 0.201 | 0.31 | 56.42 | 0.32 | 0.42 |
| FaLIM7 | 0.43 | 0.22 | 0.40 | 0.25 | 0.169 | 0.34 | 54.45 | 0.35 | 0.44 |
| FaLIM15 | 0.48 | 0.20 | 0.37 | 0.22 | 0.195 | 0.37 | 51.76 | 0.32 | 0.44 |
| FaLIM19 | 0.48 | 0.20 | 0.37 | 0.22 | 0.191 | 0.36 | 50.99 | 0.32 | 0.44 |
| FaLIM27 | 0.48 | 0.20 | 0.37 | 0.22 | 0.193 | 0.36 | 51.75 | 0.32 | 0.44 |
| FaLIM24 | 0.47 | 0.21 | 0.37 | 0.23 | 0.192 | 0.37 | 52.14 | 0.33 | 0.45 |
| FvLIM21 | 0.48 | 0.20 | 0.36 | 0.24 | 0.181 | 0.35 | 50.30 | 0.33 | 0.44 |
| FvLIM7 | 0.38 | 0.34 | 0.38 | 0.24 | 0.266 | 0.42 | 43.11 | 0.43 | 0.46 |
| FaLIM17 | 0.38 | 0.34 | 0.38 | 0.23 | 0.268 | 0.43 | 43.14 | 0.43 | 0.46 |
| FaLIM18 | 0.38 | 0.34 | 0.37 | 0.24 | 0.270 | 0.43 | 42.82 | 0.43 | 0.46 |
| FaLIM14 | 0.42 | 0.33 | 0.34 | 0.25 | 0.262 | 0.42 | 43.15 | 0.43 | 0.46 |
| FaLIM28 | 0.41 | 0.33 | 0.34 | 0.25 | 0.260 | 0.43 | 44.11 | 0.44 | 0.46 |
| FaLIM25 | 0.41 | 0.32 | 0.37 | 0.24 | 0.273 | 0.44 | 41.34 | 0.42 | 0.46 |
| FvLIM5 | 0.33 | 0.38 | 0.35 | 0.28 | 0.271 | 0.45 | 52.09 | 0.49 | 0.48 |
| 基因Gene | T3s | C3s | A3s | G3s | CAI | Fop | ENC | GC3s | GC |
| FaLIM10 | 0.32 | 0.39 | 0.34 | 0.28 | 0.273 | 0.46 | 60.52 | 0.50 | 0.48 |
| FaLIM12 | 0.32 | 0.39 | 0.34 | 0.28 | 0.272 | 0.45 | 55.80 | 0.50 | 0.48 |
| FaLIM9 | 0.32 | 0.38 | 0.35 | 0.29 | 0.265 | 0.44 | 51.91 | 0.50 | 0.47 |
| FaLIM22 | 0.39 | 0.32 | 0.29 | 0.32 | 0.334 | 0.45 | 56.12 | 0.48 | 0.47 |
| FaLIM23 | 0.39 | 0.31 | 0.28 | 0.33 | 0.348 | 0.47 | 52.97 | 0.48 | 0.47 |
| FaLIM20 | 0.39 | 0.31 | 0.29 | 0.32 | 0.333 | 0.45 | 55.85 | 0.48 | 0.47 |
| FvLIM9 | 0.39 | 0.32 | 0.29 | 0.32 | 0.332 | 0.44 | 55.51 | 0.48 | 0.47 |
| FaLIM26 | 0.39 | 0.32 | 0.29 | 0.32 | 0.332 | 0.44 | 55.51 | 0.48 | 0.47 |
| FaLIM16 | 0.39 | 0.31 | 0.29 | 0.32 | 0.351 | 0.46 | 52.26 | 0.47 | 0.47 |
| FvLIM2 | 0.29 | 0.42 | 0.34 | 0.30 | 0.266 | 0.46 | 58.30 | 0.53 | 0.47 |
| FaLIM8 | 0.27 | 0.44 | 0.34 | 0.31 | 0.242 | 0.44 | 59.77 | 0.55 | 0.48 |
| FvLIM4 | 0.29 | 0.41 | 0.38 | 0.24 | 0.254 | 0.48 | 51.73 | 0.50 | 0.47 |
| FaLIM5 | 0.29 | 0.42 | 0.38 | 0.23 | 0.259 | 0.48 | 51.62 | 0.50 | 0.47 |
| FvLIM6 | 0.40 | 0.35 | 0.28 | 0.33 | 0.318 | 0.48 | 59.48 | 0.50 | 0.45 |
| FaLIM13 | 0.41 | 0.34 | 0.29 | 0.32 | 0.316 | 0.47 | 56.22 | 0.48 | 0.45 |
| FaLIM11 | 0.40 | 0.36 | 0.28 | 0.31 | 0.320 | 0.48 | 55.92 | 0.49 | 0.45 |
| 平均 Average | 0.41 | 0.29 | 0.35 | 0.26 | 0.256 | 0.42 | 52.10 | 0.42 | 0.45 |
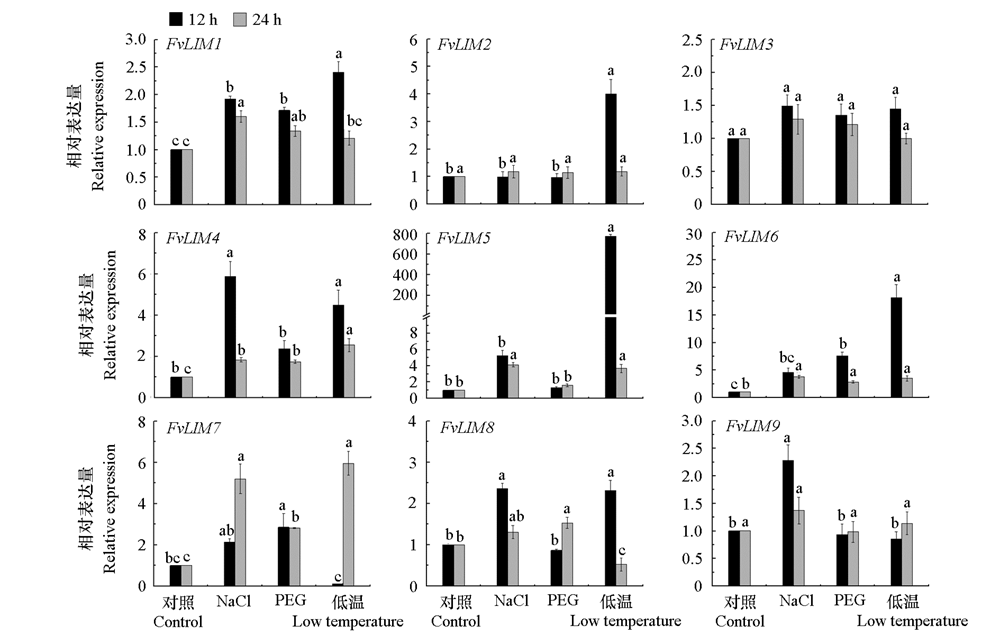
图9 森林草莓LIM家族候选基因荧光定量表达分析 不同小写字母表示处理与对照间差异显著。
Fig. 9 Quantitative expression of LIM family candidate genes in Fragaria vesca Different lowercase letters indicate that there are significant differences between the treatment and the control.
| [1] |
Agulnick A D, Taira M, Breen J J, Tanaka T, Dawid I B, Westphal H. 1996. Interactions of the LIM-domain-binding factor Ldb 1 with LIM homeodomain proteins. Nature, 384 (6606):270-272.
doi: 10.1038/384270a0 URL |
| [2] | Arnaud D, Dejardin A, Leple J C, Lesage-Descauses M C, Boizot N, Villar M, Benedetti H, Pilate G. 2012. Expression analysis of LIM gene family in poplar,toward an updated phylogenetic classifification. BMC Research, 5 (1):102. |
| [3] |
Arnaud D, Déjardin A, Leplé J C, Lesage-descauses M C, Pilate G. 2007. Genome-wide analysis of LIM gene family in Populus trichocarpa,Arabidopsis thaliana,and Oryza sativa. DNA Research, 14 (3):103-116.
doi: 10.1093/dnares/dsm013 URL |
| [4] | Bailey T L, Boden M, Buske F A, Frith M, Grant C E, Clementi L, Ren J Y, LI W W, Noble W S. 2009. MEME SUITE:tools for motif discovery and searching. Nucleic Acids Research, 37 (Web Server issue):W202. |
| [5] |
Cheng X, Li G H, Muhammad A, Zhang J Y, Jiang T S, Jin Q, Zhao H, Cai Y P, Lin Y. 2019. Molecular identification,phylogenomic characterization and expression patterns analysis of the LIM(LIN-11,Isl1 and MEC-3 domains)gene family in pear(Pyrus bretschneideri)reveal its potential role in lignin metabolism. Gene, 686:237-249.
doi: S0378-1119(18)31207-1 pmid: 30468911 |
| [6] |
Chenna R, Sugawara H, Koike T, Lopez R, Gibson T J, Higgins D G, Thompson J D. 2003. Multiple sequence alignment with the clustal series of programs. Nucleic Acids Research, 31 (13):3497-3500.
pmid: 12824352 |
| [7] |
Dixon R A, Paiva N L. 1995. Stress-induced phenylpropanoid metabolism. The Plant Cell, 7 (7):1085-1097.
pmid: 12242399 |
| [8] |
Feng L, Xia R, Liu Y L. 2019. Comprehensive characterization of miRNA and PHAS loci in the diploid strawberry(Fragaria vesca)genome. Horticultural Plant Journal, 5 (6):255-267.
doi: 10.1016/j.hpj.2019.11.004 |
| [9] | Gao Ya. 2019. Genetic transformation of Gossypium hirsutum and function of LIM gene in offspring of transgenic cotton[M. D. Dissertation]. Wuhan: Central China Normal University. (in Chinese) |
| 高雅. 2019. 棉花(Gossypium hirsutum)遗传转化及LIM基因在转基因棉花后代中的功能研究[硕士论文]. 武汉: 华中师范大学. | |
| [10] | He Hong-hong, Ma Zong-huan, Zhang Yuan-xia, Zhang Juan, Lu Shi-xiong, Zhang Zhi-qiang, Zhao Xin, Wu Yu-xia, Mao Juan. 2018. Identification and expression analysis of LBD gene family in grape. Scientia Agricultura Sinica, 51 (21):4102-4118. (in Chinese) |
| 何红红, 马宗桓, 张元霞, 张娟, 卢世雄, 张志强, 赵鑫, 吴玉霞, 毛娟. 2018. 葡萄LBD基因家族的鉴定与表达分析. 中国农业科学, 51 (21):4102-4118. | |
| [11] |
Jurata L W, Gill G N. 1997. Functional analysis of the nuclear LIM domain interactor NLI. Molecular and Cellular Biology, 17 (10):5688-5698.
doi: 10.1128/MCB.17.10.5688 pmid: 9315627 |
| [12] | Kadrmas J L, Beckerle M C. 2004. The LIM domain:from the cytoskeleton to the nucleus. Nature Reviews. Molecular Cell Biology, 5 (11):920-931. |
| [13] |
Kawaoka A, Ebinuma H. 2001. Transcriptional control of lignin biosynthesis by tobacco LIM protein. Phytochemistry, 57 (7):1149-1157.
pmid: 11430987 |
| [14] |
Khadiza K, Arif H K R, Jong-In P, Nasar U A, Chang K K, Ki-Byung L, Min-Bae K, Do-Jin L, Ill S N, Mi-Young C. 2016. Genome-wide identification,characterization and expression profiling of LIM family genes in Solanum lycopersicum L. Plant Physiology and Biochemistry, 108:177-190.
doi: S0981-9428(16)30273-X pmid: 27439220 |
| [15] | Li Tong. 2017. Transcriptional regulation of sorghum SbLIM1 on lignin synthesis and recognition of interacting proteins[M. D. Dissertation]. Jinan: Shandong University. (in Chinese) |
| 李彤. 2017. 高梁SbLIM1对木质素合成的转录调控及互作蛋白识别研究[硕士论文]. 济南:山东大学. | |
| [16] |
Li Ya, Yue Xiao-feng, Que Ya-wei, Yan Xia, Ma Zhong-hua, Talbot Nicholas J, Wang Zheng-yi. 2014. Characterisation of four LIM protein-encoding genes involved in infection-related development and pathogenicity by the rice blast fungus Magnaporthe oryzae. PLoS One, 9 (2):e88246.
doi: 10.1371/journal.pone.0088246 URL |
| [17] | Lian Peng, Du Meng-qing, Wang Li-juan. 2019. Tissue culture and disinfection of strawberry stem tips and optimization of hormone ratio. Journal of Tianjin Agricultural College, 26 (3):21-25. (in Chinese) |
| 连朋, 杜梦卿, 王丽娟. 2019. 草莓茎尖组培消毒方法及激素配比的优选. 天津农学院学报, 26 (3):21-25. | |
| [18] | Liu Tao, Wang Ping-ping, He Hong-hong, Liang Guo-ping, Lu Shi-xiong, Chen Bai-hong, Mao Juan. 2020. Identification and expression analysis of CIPK gene family in strawberry. Acta Horticulturae Sinica, 47 (1):127-142. (in Chinese) |
| 刘涛, 王萍萍, 何红红, 梁国平, 卢世雄, 陈佰鸿, 毛娟. 2020. 草莓CIPK基因家族的鉴定与表达分析. 园艺学报, 47 (1):127-142. | |
| [19] | Liu Wan-da, Zhang Bing-xiu, Wei Yuan-yuan, Wang Yu, Dong Chao, Zhu Yan-ming. 2014. Analysis of codon preference of strawberry DHAR gene. Northern Horticulture,(14):92-97. (in Chinese) |
| 刘万达, 张丙秀, 魏媛媛, 王禹, 董超, 朱延明. 2014. 草莓DHAR基因密码子偏好性分析. 北方园艺,(14):92-97. | |
| [20] | Liu Zhi. 2011. Functional analysis of Oslim transcription factor involved in non-host resistance of rice[M. D. Dissertation]. Beijing:Chinese Academy of Agricultural Sciences. (in Chinese) |
| 刘峙. 2011. LIM转录因子(Oslim)参与水稻非寄主抗性的功能分析[硕士论文]. 北京:中国农业科学院. | |
| [21] |
Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real-time quantitative and the 2-ΔΔCT method. Methods, 25 (4):402-408.
pmid: 11846609 |
| [22] | Ma Zong-huan, Mao Juan, Li Wen-fang, Yang Shi-mao, Wu Jin-hong, Chen Bai-hong. 2016. Identification and expression analysis of SnRK 2 family genes in grape. Acta Horticulturae Sinica, 43 (10):1891-1903. (in Chinese) |
| 马宗桓, 毛娟, 李文芳, 杨世茂, 吴金红, 陈佰鸿. 2016. 葡萄SnRK2家族基因的鉴定与表达分析. 园艺学报, 43 (10):1891-1903. | |
| [23] | Mao Juan, Lu Shi-xiong, Wang Ping, Ma Zong-huan, Liu Tao, He Hong-hong, Nai Guo-jie, Zhang Zhi-qiang, Chen Bai-hong. 2020. Analysis of gene characteristic and expression pattern of LIM family in grape. Acta Horticulturae Sinica, 47 (7):1277-1288. (in Chinese) |
| 毛娟, 卢世雄, 王萍, 马宗桓, 刘涛, 何红红, 乃国洁, 张志强, 陈佰鸿. 2020. 葡萄LIM家族基因特征及表达模式分析. 园艺学报, 47 (7):1277-1288. | |
| [24] | Michelsen J W, Schmeichel K L, Beckerle M C, Winge D R. 1993. The LIM motif defines a specific zinc-binding protein domain. Proceedings of the National Academy of Sciences of the United States of America, 90 (10):4404-4408. |
| [25] |
Rosane L C, Ellen C P, Isabel L V, Rafael W, Amilton S, Luciano D S P, César V R, Vanessa G. 2020. Genome-wide identification,and characterization of the CDPK gene family reveal their involvement in abiotic stress response in Fragaria ananassa. Scientific Reports, 10 (1):1-17.
doi: 10.1038/s41598-019-56847-4 URL |
| [26] |
Sánchez-García I, Rabbitts T H. 1994. The LIM domain:a new structural motif found in zinc-finger-like proteins. Trends in Genetics, 10 (9):315-320.
pmid: 7974745 |
| [27] | Schmeichel K L, Beckerle M C. 1994. The LIM domain is a modular protein-binding interface. MC Beckerle-Cell, 79 (2):211-219. |
| [28] |
Schmeichel K L, Beckerle M C. 1997. Molecular dissection of a LIM domain. Molecular Biology of the Cell, 8 (2):219-230.
pmid: 9190203 |
| [29] |
Taira M, Jamrich M, Good P J, Dawid I B. 1992. The LIM domain-containing homeo box gene Xlim-1 is expressed specifically in the organizer region of Xenopus gastrula embryos. Genes & Development, 6 (3):356-366.
doi: 10.1101/gad.6.3.356 URL |
| [30] | Wang Yong-qiang, He Xiao-liang, Liu Jian-guang, Du Shu-feng, Zhao Jun-li, Zhao Gui-yuan, Zhang Han-shan. 2014. Cloning and expression analysis of GhLIMa in cotton salt-tolerant gene. Acta Botanica Boreali-Occidentalis, 34 (12):2382-2387. (in Chinese) |
| 王永强, 何晓亮, 刘建光, 杜淑凤, 赵俊丽, 赵贵元, 张寒霜. 2014. 棉花耐盐相关基因GhLIMa的克隆与表达分析. 西北植物学报, 34 (12):2382-2387. | |
| [31] |
Wittkopp P J, Kalay G. 2011. Cis-regulatory elements:molecular mechanisms and evolutionary processes underlying divergence. Nature Reviews Genetics, 13 (1):59-69.
doi: 10.1038/nrg3095 URL |
| [32] | Xu Yang, Yuan Li-ping, Chen Jin-ling, Huang Xiao-hong. 2012. Research progress of actin depolymerizing factor/Cofilin function. Life Sciences, 24 (1):13-18. (in Chinese) |
| 徐阳, 原丽平, 陈金铃, 黄晓红. 2012. 肌动蛋白解聚因子/cofilin功能的研究进展. 生命科学, 24 (1):13-18. | |
| [33] |
Yang R, Chen M, Sun J C, Yu Y, Min D H, Chen J, Xu Z S, Zhou Y B, Ma Y Z, Zhang X H. 2019. Genome-wide analysis of LIM family genes in foxtail millet(Setaria italica L.)and characterization of the role of SiWLIM2b in drought tolerance. International Journal of Molecular Sciences, 20 (6):1303.
doi: 10.3390/ijms20061303 URL |
| [34] | Yang Rui. 2019. Effects of SiWLIM2b gene on drought resistance and low nitrogen stress tolerance in rice[M. D. Dissertation]. Xianyang:Northwest A & F University. (in Chinese) |
| 杨瑞. 2019. 谷子SiWLIM2b基因提高水稻抗旱及耐低氮胁迫的功能研究[硕士论文]. 咸阳: 西北农林科技大学. | |
| [35] | Li Bo, Hu wen-ran, Zhang Jing-bo, Fan Ling. 2015. Evolution and expression analysis of the LIM protein gene family in cotton. Plant Physiology Journal, 12:2133-2142. (in Chinese) |
| 杨洋, 李波, 胡文冉, 张经博, 范玲. 2015. 棉花LIM蛋白基因家族的进化及表达特征分析. 植物生理学报, 12:2133-2142. | |
| [36] |
Ye J R, Zhou L M, Xu M L. 2013. Arabidopsis LIM proteins PLIM2a and PLIM2b regulate actin configuration during pollen tube growth. Biologia Plantarum, 57 (3):433-441.
doi: 10.1007/s10535-013-0323-3 URL |
| [37] | Yi F, Guo J, Dabbagh D, Spear M, He S J, Kehn-Hall K, Fontenot J, Yin Y, Bibian M, Park C M, Zheng K, Park H J, Soloveva V, Gharaibeh D, Retterer C, Zamani R, Naughton J, Jiang Y J, Shang H, Hakami R M, Ling B H, Young J A T, Bavari S, Xu X H, Feng Y B, Wu Y T. 2017. Discovery of novel small-molecule inhibitors of LIM domain kinase for inhibiting HIV-1. Journal of Virology, 91 (13):e02418-16. |
| [38] | Yuan Gao-peng, Han Xiao-lei, Bian Shu-xun, Zhang Li-yi, Tian Yi, Zhang Cai-xia, Cong Peihua. 2019. Bioinformatics and expression analysis of apple LIM gene family. Scientia Agricultura Sinica, 52 (23):4322-4332. (in Chinese) |
| 袁高鹏, 韩晓蕾, 卞书迅, 张利义, 田义, 张彩霞, 丛佩华. 2019. 苹果LIM基因家族生物信息学及表达分析. 中国农业科学, 52 (23):4322-4332. | |
| [39] | Zhang Hai-yan, Li Zuo-tong, Zhao Chang-jiang, Yang Ke-jun, Wang Yu-feng, Hu Xue-wei, Zhao Ying. 2013. Analysis of LIM domain protein gene family in maize. Maize Science, 21 (3):40-47. (in Chinese) |
| 张海燕, 李佐同, 赵长江, 杨克军, 王玉凤, 胡雪微, 赵莹. 2013. 玉米LIM结构域蛋白基因家族分析. 玉米科学, 21 (3):40-47. | |
| [40] |
Zhang X, Zhen J B, Li Z H, Kang D M, Yang Y M, Kong J, Hua J P. 2011. Expression profile of early responsive genes under salt stress in upland cotton ( Gossypium hirsutum L.). Plant Molecular Biology Reporter, 29 (3):626-637.
doi: 10.1007/s11105-010-0269-y URL |
| [41] | Zhang Yi-han, LI Qing, Zhang Lei, Tan He-xin, Chen Xiang-hui. 2019. Bioinformatics analysis of the MADs-Box gene family in Crocus sativus. Genomics and Applied Biology, 38 (7):3140-3154. (in Chinese) |
| 张一菡, 李卿, 张磊, 谭何新, 陈祥慧. 2019. 西红花MADS-box基因家族的生物信息学分析. 基因组学与应用生物学, 38 (7):3140-3154. | |
| [42] |
Zhang Z B, Zhang J W, Chen Y J, Li R F, Wang H Z, Wei J H. 2012. Genome-wide analysis and identification of HAK potassium transporter gene family in maize(Zea mays L.). Molecular Biology Reports, 39 (8):8465-8473.
doi: 10.1007/s11033-012-1700-2 URL |
| [43] | Zhou Jian-ping. 2006. Phenotypic changes in cotton induced by excessive expression of LIM domain gene[M. D. Dissertation]. Chongqing:Southwest University. (in Chinese) |
| 周建平. 2006. 超量表达LIM结构域基因引起棉花的表型变化[硕士论文]. 重庆:西南大学. |
| [1] | 宋艳红, 陈亚铎, 张晓玉, 宋 盼, 刘丽锋, 李 刚, 赵 霞, 周厚成, . 森林草莓FvbHLH130转录因子调控植株提前开花[J]. 园艺学报, 2023, 50(2): 295-306. |
| [2] | 何成勇, 赵晓丽, 许腾飞, 高德航, 李世访, 王红清. 草莓病毒1山东分离物全基因组分析[J]. 园艺学报, 2023, 50(1): 153-160. |
| [3] | 杨 雷, 李 莉, 董 辉, 冯 佳, 张建军, 范婧芳, 杨秋叶, 杨 莉, . 草莓新品种‘石莓11号’[J]. 园艺学报, 2022, 49(S2): 79-80. |
| [4] | 赵 霞, 李 刚, 刘丽锋, 宋艳红, 周厚成. 草莓新品种‘华硕1号’[J]. 园艺学报, 2022, 49(S2): 81-82. |
| [5] | 董 静, 常琳琳, 王桂霞, 钟传飞, 隗永青, 孙 健, 孙 瑞, 张宏力, 高用顺, 许利平, 陶 磅, 罗志伟, 张运涛, . 四季草莓新品种‘静红’[J]. 园艺学报, 2022, 49(S1): 61-62. |
| [6] | 蔡建法, 莫雪莲, 管思聪, 陈栩, 薛程. 草莓FvYABBY5.1表达特性和功能分析[J]. 园艺学报, 2022, 49(7): 1458-1472. |
| [7] | 钱婕妤, 蒋玲莉, 郑钢, 陈佳红, 赖吴浩, 许梦晗, 付建新, 张超. 百日草花青素苷合成相关MYB转录因子筛选及ZeMYB9功能研究[J]. 园艺学报, 2022, 49(7): 1505-1518. |
| [8] | 王宏, 杨王莉, 蔺经, 杨青松, 李晓刚, 盛宝龙, 常有宏. 早熟砂梨‘苏翠1号’与其亲本‘翠冠’‘华酥’成熟果实差异代谢产物及差异基因比较分析[J]. 园艺学报, 2022, 49(3): 493-508. |
| [9] | 姜翠翠, 方智振, 周丹蓉, 林炎娟, 叶新福. ‘芙蓉李’糖转运蛋白家族基因鉴定及表达分析[J]. 园艺学报, 2022, 49(2): 252-264. |
| [10] | 王光鹏, 刘同坤, 徐新凤, 李竹帛, 高瞻远, 侯喜林. 大白菜LEA家族基因的鉴定及其部分成员在低温胁迫下的表达分析[J]. 园艺学报, 2022, 49(2): 304-318. |
| [11] | 杨丽媛, 王倩, 王许会, 徐通达, 马军. 草莓生长素合成关键酶FveTAA1保守氨基酸位点T111的生物学功能研究[J]. 园艺学报, 2021, 48(9): 1695-1705. |
| [12] | 洪燕红, 叶清华, 李泽坤, 王威, 谢倩, 陈清西, 陈建清. 红花草莓‘莓红’花瓣花色苷积累及其MYB基因的表达分析[J]. 园艺学报, 2021, 48(8): 1470-1484. |
| [13] | 黄倩茹, 牛永浩, 吴宽, 刘占杰, 曹孟籍, 赵磊, 吴云锋. 陕西省草莓病毒种类的LncRNA测序初步鉴定[J]. 园艺学报, 2021, 48(8): 1589-1594. |
| [14] | 朱俊飞, 李鑫, 董康挺, 唐志菲, 边秀举, 王丽宏, 李会彬, 孙鑫博. 转匍匐翦股颖AsHSP26.8a拟南芥株系的光合作用研究[J]. 园艺学报, 2021, 48(8): 1619-1625. |
| [15] | 谷思, 刘璐, 李安然, 张伟伟, 赵帅琪, 邢宇. 草莓果实酵母双杂交文库的构建及FvM4K1互作蛋白的筛选[J]. 园艺学报, 2021, 48(6): 1067-1078. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司