
园艺学报 ›› 2021, Vol. 48 ›› Issue (7): 1317-1328.doi: 10.16420/j.issn.0513-353x.2020-0656
杨双娟, 张晓伟, 魏小春, 赵艳艳, 王志勇, 赵肖斌, 李林, 原玉香**( )
)
收稿日期:2020-12-24
修回日期:2021-02-19
出版日期:2021-07-25
发布日期:2021-08-10
通讯作者:
原玉香
E-mail:yuanyuxiang@126.com
基金资助:
YANG Shuangjuan, Zhang Xiaowei, WEI Xiaochun, ZHAO Yanyan, WANG Zhiyong, ZHAO Xiaobin, LI Lin, YUAN Yuxiang**( )
)
Received:2020-12-24
Revised:2021-02-19
Online:2021-07-25
Published:2021-08-10
Contact:
YUAN Yuxiang
E-mail:yuanyuxiang@126.com
摘要:
以大白菜1E519BC1F2和4E511BC1F2两个群体为试材,分别构建根肿病抗病和感病池,进行集团分离群体测序(Bulked Segregant Analysis Sequencing,BSA-seq)和分析。结果发现,两个群体中检测到1个共同的根肿病抗性基因候选区间,位于A08染色体7.40 ~ 8.85 Mb,命名为BraA.Pb.8.4。该基因在物理位置上和已经报道的根肿病抗性基因Crr1、Rcr9和CRs均不同,并且Crr1和Rcr9连锁的标记在抗病和感病材料中不具有多态性,CRs连锁的4个标记中只有1个标记(Probe60)在抗病和感病材料中存在多态性,表明BraA.Pb.8.4不同于已经报道的基因,可能为1个新的根肿病抗性基因。在BraA.Pb.8.4候选区间开发了3个KASP标记(BraA.Pb.8.4-K1、BraA.Pb.8.4-K2和BraA.Pb.8.4-K3),均可以有效将纯合抗病、纯合感病和杂合抗病材料区分开,为共显性标记。利用标记BraA.Pb.8.4-K1在5个群体共257个单株中进行验证,发现各单株的基因型和抗病性表型平均一致率高达96.13%。因此,本研究鉴定的BraA.Pb.8.4基因及开发的KASP标记可以高效用于根肿病的分子标记辅助育种。
中图分类号:
杨双娟, 张晓伟, 魏小春, 赵艳艳, 王志勇, 赵肖斌, 李林, 原玉香. 大白菜抗根肿病基因BraA.Pb.8.4的定位及KASP标记开发[J]. 园艺学报, 2021, 48(7): 1317-1328.
YANG Shuangjuan, Zhang Xiaowei, WEI Xiaochun, ZHAO Yanyan, WANG Zhiyong, ZHAO Xiaobin, LI Lin, YUAN Yuxiang. Mapping and KASP Markers Development for Clubroot Resistance Gene BraA.Pb.8.4 in Chinese Cabbage[J]. Acta Horticulturae Sinica, 2021, 48(7): 1317-1328.
| 样品名称 Sample | 高质量读长数 Clean reads | 高质量碱基数 Clean bases | 比对的读长数 Mapped reads | 比对率/% Mapping rate | 基因组覆盖倍数 Coverage of B. rapa genome |
|---|---|---|---|---|---|
| 1E519BC1F2-Rp | 146 359 698 | 21 953 954 700 | 139 582 489 | 95.37 | 45.27 |
| 1E519BC1F2-Sp | 130 400 096 | 19 560 014 400 | 124 262 099 | 95.29 | 40.33 |
| 4E511BC1F2-Rp | 171 165 594 | 25 674 839 100 | 163 000 999 | 95.23 | 52.94 |
| 4E511BC1F2-Sp | 125 843 472 | 18 876 520 800 | 119 448 979 | 94.92 | 38.92 |
表1 测序数据统计
Table 1 Summary of whole-genome re-sequencing data
| 样品名称 Sample | 高质量读长数 Clean reads | 高质量碱基数 Clean bases | 比对的读长数 Mapped reads | 比对率/% Mapping rate | 基因组覆盖倍数 Coverage of B. rapa genome |
|---|---|---|---|---|---|
| 1E519BC1F2-Rp | 146 359 698 | 21 953 954 700 | 139 582 489 | 95.37 | 45.27 |
| 1E519BC1F2-Sp | 130 400 096 | 19 560 014 400 | 124 262 099 | 95.29 | 40.33 |
| 4E511BC1F2-Rp | 171 165 594 | 25 674 839 100 | 163 000 999 | 95.23 | 52.94 |
| 4E511BC1F2-Sp | 125 843 472 | 18 876 520 800 | 119 448 979 | 94.92 | 38.92 |
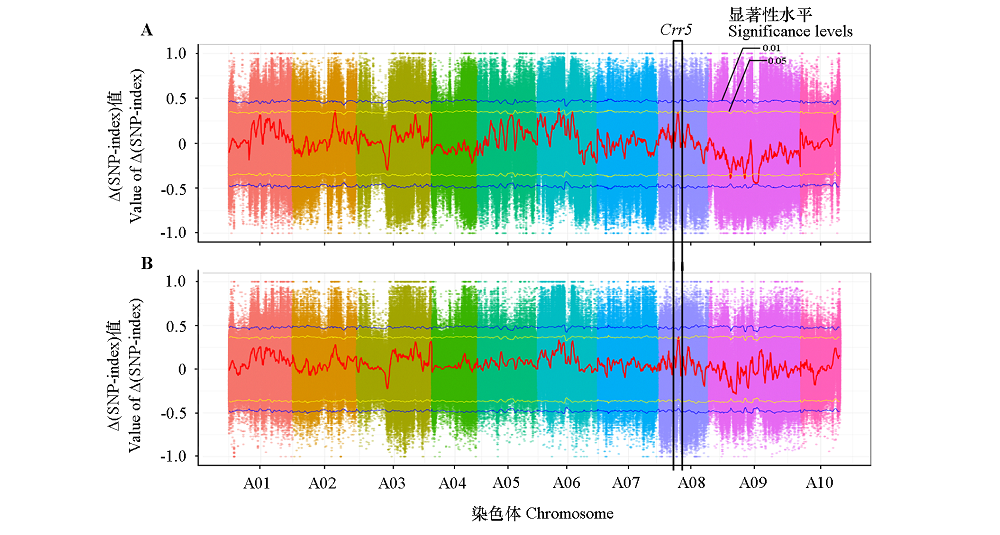
图3 大白菜1E519BC1F2(A)和4E511BC1F2(B)群体的BSA-seq分析结果 两个群体共同的候选区间命名为BraA.Pb.8.4;曼哈顿图中黄色线为0.05显著水平,蓝色线为0.01显著水平。
Fig. 3 BSA-seq analysis of 1E519BC1F2(A)and 4E511BC1F2(B)populations in Chinese cabbage The common candidate region in the two populations is named as BraA.Pb.8.4;the yellow and blue line in Manhattan plots indicate 0.05 and 0.01 significance levels,respectively.
| 群体 Population | 位置/Mb Location | 抗病混池SNP index Rp_SNP_index | 感病混池SNP index Sp_SNP_index | ∆SNP-index | P值 P-value |
|---|---|---|---|---|---|
| 1E519BC1F2 | A08:7.55 ~ 8.75 | 0.48 | 0.14 | 0.34 | < 0.05 |
| 4E511BC1F2 | A08:7.40 ~ 8.85 | 0.45 | 0.10 | 0.35 | < 0.05 |
表2 两个群体的共同定位区间信息
Table 2 Information of the common candidate region in two populations
| 群体 Population | 位置/Mb Location | 抗病混池SNP index Rp_SNP_index | 感病混池SNP index Sp_SNP_index | ∆SNP-index | P值 P-value |
|---|---|---|---|---|---|
| 1E519BC1F2 | A08:7.55 ~ 8.75 | 0.48 | 0.14 | 0.34 | < 0.05 |
| 4E511BC1F2 | A08:7.40 ~ 8.85 | 0.45 | 0.10 | 0.35 | < 0.05 |
| KASP标记 KASP marker | 物理位置 Physical position | 变异位点 Variation locus | 引物序列(5′-3′) Primer sequence |
|---|---|---|---|
| BraA.Pb.8.4-K1 | A08:7 953 439 | T/C | Fa:GAAGGTGACCAAGTTCATGCTCGTGGTGGAAAGAAATGGTT |
| Fb:GAAGGTCGGAGTCAACGGATTCGTGGTGGAAAGAAATGGTC | |||
| R:CGTCCTCCGAGAAGCACACACC | |||
| BraA.Pb.8.4-K2 | A08:7 961 681 | T/G | Fa:GAAGGTCGGAGTCAACGGATTCGTCCTGCATTTCTTAGATGGT |
| Fb:GAAGGTGACCAAGTTCATGCTCGTCCTGCATTTCTTAGATGGG | |||
| R:CGAGCACTGATCTGCAAACACA | |||
| BraA.Pb.8.4-K3 | A08:7 970 088 | C/T | Fa:GAAGGTGACCAAGTTCATGCTGTTTAAGTAGAGACTGTGGGAC |
| Fb:GAAGGTCGGAGTCAACGGATTGTTTAAGTAGAGACTGTGGGAT | |||
| R:CTTCAAACGCAGCTTTCTTTGACTT |
表3 基因BraA.Pb.8.4的KASP标记信息
Table 3 Information of KASP markers linked to gene BraA.Pb.8.4
| KASP标记 KASP marker | 物理位置 Physical position | 变异位点 Variation locus | 引物序列(5′-3′) Primer sequence |
|---|---|---|---|
| BraA.Pb.8.4-K1 | A08:7 953 439 | T/C | Fa:GAAGGTGACCAAGTTCATGCTCGTGGTGGAAAGAAATGGTT |
| Fb:GAAGGTCGGAGTCAACGGATTCGTGGTGGAAAGAAATGGTC | |||
| R:CGTCCTCCGAGAAGCACACACC | |||
| BraA.Pb.8.4-K2 | A08:7 961 681 | T/G | Fa:GAAGGTCGGAGTCAACGGATTCGTCCTGCATTTCTTAGATGGT |
| Fb:GAAGGTGACCAAGTTCATGCTCGTCCTGCATTTCTTAGATGGG | |||
| R:CGAGCACTGATCTGCAAACACA | |||
| BraA.Pb.8.4-K3 | A08:7 970 088 | C/T | Fa:GAAGGTGACCAAGTTCATGCTGTTTAAGTAGAGACTGTGGGAC |
| Fb:GAAGGTCGGAGTCAACGGATTGTTTAAGTAGAGACTGTGGGAT | |||
| R:CTTCAAACGCAGCTTTCTTTGACTT |

图4 利用标记BraA.Pb.8.4-K1(A、B)和BraA.Pb.8.4-K2(C、D)对1E519BC1F2群体进行KASP基因分型 A、C:KASP基因分型结果;B、D:X-Y坐标轴下KASP基因分型结果。
Fig. 4 KASP genotyping of 1E519BC1F2 population with primers BraA.Pb.8.4-K1(A,B)and BraA.Pb.8.4-K2(C,D) A,C:KASP genotyping result in 96-well plate format;B,D:KASP genotyping result in X-Y coordinates.
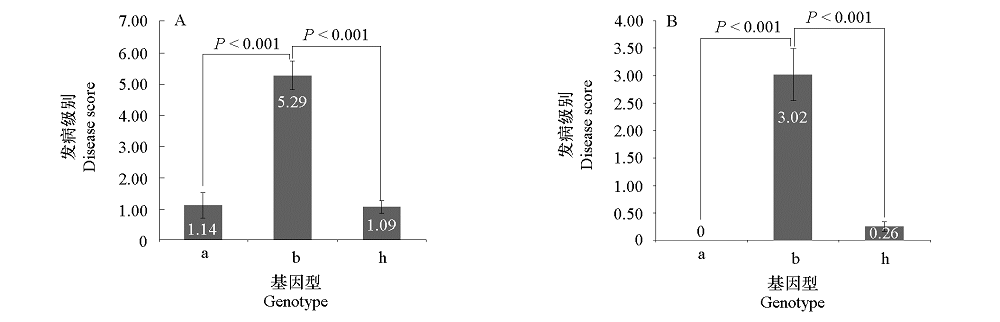
图5 利用1E519BC1F2(A)和4E511BC1F2(B)群体验证标记BraA.Pb.8.4-K1基因型和根肿病抗性间的相关性
Fig. 5 Correlation analysis between genotypes of marker BraA.Pb.8.4-K1 and clubroot resistance phenotypes in population 1E519BC1F2(A)and 4E511BC1F2(B)
| 群体 Population | 根肿病抗性 Clubroot resistance | 单株数 Number of individual | 基因型 Genotype | 一致率/% Concordance rate | ||
|---|---|---|---|---|---|---|
| a | b | h | ||||
| 1E177BC2F2-5 | 抗病Resistant | 19 | 0 | 1 | 18 | 94.74 |
| 感病Susceptible | 19 | 0 | 18 | 1 | ||
| 1E177BC2F2-24 | 抗病Resistant | 28 | 9 | 0 | 19 | 100.00 |
| 感病Susceptible | 11 | 0 | 11 | 0 | ||
| 1E177BC2F2-25 | 抗病Resistant | 34 | 9 | 2 | 23 | 91.30 |
| 感病Susceptible | 12 | 0 | 10 | 2 | ||
| 1E177BC2F2-26 | 抗病Resistant | 29 | 6 | 0 | 23 | 100.00 |
| 感病Susceptible | 12 | 0 | 12 | 0 | ||
| CR1E-10 | 抗病Resistant | 45 | 0 | 5 | 40 | 94.62 |
| 感病Susceptible | 48 | 0 | 48 | 0 | ||
表4 标记BraA.Pb.8.4-K1在5个群体中的验证
Table 4 Validation of marker BraA.Pb.8.4-K1 in five populations
| 群体 Population | 根肿病抗性 Clubroot resistance | 单株数 Number of individual | 基因型 Genotype | 一致率/% Concordance rate | ||
|---|---|---|---|---|---|---|
| a | b | h | ||||
| 1E177BC2F2-5 | 抗病Resistant | 19 | 0 | 1 | 18 | 94.74 |
| 感病Susceptible | 19 | 0 | 18 | 1 | ||
| 1E177BC2F2-24 | 抗病Resistant | 28 | 9 | 0 | 19 | 100.00 |
| 感病Susceptible | 11 | 0 | 11 | 0 | ||
| 1E177BC2F2-25 | 抗病Resistant | 34 | 9 | 2 | 23 | 91.30 |
| 感病Susceptible | 12 | 0 | 10 | 2 | ||
| 1E177BC2F2-26 | 抗病Resistant | 29 | 6 | 0 | 23 | 100.00 |
| 感病Susceptible | 12 | 0 | 12 | 0 | ||
| CR1E-10 | 抗病Resistant | 45 | 0 | 5 | 40 | 94.62 |
| 感病Susceptible | 48 | 0 | 48 | 0 | ||
| 基因 Gene | 标记或候选基因 Marker or candidate gene | A08物理位置/bp Physical position on A08 |
|---|---|---|
| BraA.Pb.8.4 | BraA.Pb.8.4-K1 | 7 953 439 |
| BraA.Pb.8.4-K2 | 7 961 681 | |
| BraA.Pb.8.4-K3 | 7 970 088 | |
| CRs | Probe55 | 9 956 858 |
| Probe59 | 10 497 705 | |
| Probe60 | 10 539 495 | |
| Probe64 | 10 707 395 | |
| Rcr9 | Bra020936 | 10 295 068 ~ 10 295 637 |
| Crr1 | Bra020861 | 10 809 433 ~ 10 825 238 |
表5 BraA.Pb.8.4和其他A08染色体上基因物理位置的比较
Table 5 Comparison the physical position of BraA.Pb.8.4 and other genes in chromosome A08
| 基因 Gene | 标记或候选基因 Marker or candidate gene | A08物理位置/bp Physical position on A08 |
|---|---|---|
| BraA.Pb.8.4 | BraA.Pb.8.4-K1 | 7 953 439 |
| BraA.Pb.8.4-K2 | 7 961 681 | |
| BraA.Pb.8.4-K3 | 7 970 088 | |
| CRs | Probe55 | 9 956 858 |
| Probe59 | 10 497 705 | |
| Probe60 | 10 539 495 | |
| Probe64 | 10 707 395 | |
| Rcr9 | Bra020936 | 10 295 068 ~ 10 295 637 |
| Crr1 | Bra020861 | 10 809 433 ~ 10 825 238 |
| 标记 Marker | 正向引物序列(5′-3′) Forward primer | 反向引物序列(5′-3′) Reverse primer | 文献来源 Reference |
|---|---|---|---|
| Probe55 | ATTCAGATGACCTTCTTCCGAC | GATATGTTACCACGGCGAAAAC | Laila et al.,2019 |
| Probe59 | ACAGCAAAACCTCTCTTCTCTC | ATGTCAAACAAGCGTCTAAAGC | Laila et al.,2019 |
| Probe60 | ACACTTAGGTGAAGAAGCAACA | GCATGCATCAAGTAACTTGCAA | Laila et al.,2019 |
| Probe 64 | GTTGGAGAACACACTGAGATCA | TCTCTCGAGGCTTCTGTTACTA | Laila et al.,2019 |
| Rcr9_specific | AGTGGAGAACCAAAGCCAAA | CAATTGTGGCCACCTTCTTC | Yu et al.,2017 |
| BAS2 | TGTTAGACATCAAAGAGGGCTTGAG | TCTCAAAGACGATAATGAACCCAAA | Suwabe et al.,2012 |
| BAS7 | AAGCAGAAGGGTTCTTGTCTGAACT | TCACTCTGAGACATGAGGACACTTG | Suwabe et al.,2012 |
表6 BraA.Pb.8.4新颖性验证引物
Table 6 Primers used for validation the novelty of BraA.Pb.8.4
| 标记 Marker | 正向引物序列(5′-3′) Forward primer | 反向引物序列(5′-3′) Reverse primer | 文献来源 Reference |
|---|---|---|---|
| Probe55 | ATTCAGATGACCTTCTTCCGAC | GATATGTTACCACGGCGAAAAC | Laila et al.,2019 |
| Probe59 | ACAGCAAAACCTCTCTTCTCTC | ATGTCAAACAAGCGTCTAAAGC | Laila et al.,2019 |
| Probe60 | ACACTTAGGTGAAGAAGCAACA | GCATGCATCAAGTAACTTGCAA | Laila et al.,2019 |
| Probe 64 | GTTGGAGAACACACTGAGATCA | TCTCTCGAGGCTTCTGTTACTA | Laila et al.,2019 |
| Rcr9_specific | AGTGGAGAACCAAAGCCAAA | CAATTGTGGCCACCTTCTTC | Yu et al.,2017 |
| BAS2 | TGTTAGACATCAAAGAGGGCTTGAG | TCTCAAAGACGATAATGAACCCAAA | Suwabe et al.,2012 |
| BAS7 | AAGCAGAAGGGTTCTTGTCTGAACT | TCACTCTGAGACATGAGGACACTTG | Suwabe et al.,2012 |
| [1] | Chen J, Jing J, Zhan Z, Zhang T, Zhang C, Piao Z. 2013. Identification of novel QTLs for isolate-specific partial resistance to Plasmodiophora brassicae in Brassica rapa. PLoS ONE, 8:e85307. |
| [2] |
Chu M, Song T, Falk K C, Zhang X, Liu X, Chang A, Lahlali R, McGregor L, Gossen B D, Yu F. 2014. Fine mapping of Rcr1 and analyses of its effect on transcriptome patterns during infection by Plasmodiophora brassicae. BMC Genomics, 15:1166.
doi: 10.1186/1471-2164-15-1166 URL |
| [3] |
Hatakeyama K, Suwabe K, Tomita R N, Kato T, Nunome T, Fukuoka H, Matsumoto S. 2013. Identification and characterization of Crr1a,a gene for resistance to clubroot disease(Plasmodiophora brassicae Woronin.)in Brassica rapa L. PLoS ONE, 8:e54745.
doi: 10.1371/journal.pone.0054745 URL |
| [4] |
Hirai M, Harada T, Kubo N, Tsukada M, Suwabe K, Matsumoto S. 2004. A novel locus for clubroot resistance in Brassica rapa and its linkage markers. Theoretical Applied Genetics, 108:639-643.
doi: 10.1007/s00122-003-1475-x URL |
| [5] |
Laila R, Park J I, Robin A H K, Natarajan S, Vijayakumar H, Shirasawa K, Isobe S, Kim H T, Nou I S. 2019. Mapping of a novel clubroot resistance QTL using ddRAD-seq in Chinese cabbage(Brassica rapa L.). BMC Plant Biology, 19(1):13.
doi: 10.1186/s12870-018-1615-8 URL |
| [6] |
Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25:1754-1760.
doi: 10.1093/bioinformatics/btp324 URL |
| [7] |
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics, 25:2078-2079.
doi: 10.1093/bioinformatics/btp352 URL |
| [8] | Long Tong, Zhang Yani, Yang Nina, Cao Chunxia, Zhang Guangyang, Zhang Shilong, Cheng Xianliang, Huang Daye, Wan Zhongyi. 2019. Progress in biological control of Chinese cabbage clubroot disease. Hubei Agricultural Sciences, 58(5):5-8. (in Chinese) |
| 龙同, 张亚妮, 杨妮娜, 曹春霞, 张光阳, 张士龙, 程贤亮, 黄大野, 万中义. 2019. 大白菜根肿病生物防治研究进展. 湖北农业科学, 58(5):5-8. | |
| [9] |
Matsumoto E, Yasui C, Ohi M, Tsukada M. 1998. Linkage analysis of RFLP markers for clubroot resistance and pigmentation in Chinese cabbage(Brassica rapa ssp. pekinensis). Euphytica, 104:79.
doi: 10.1023/A:1018370418201 URL |
| [10] |
Pang W, Fu P, Li X, Zhan Z, Yu S, Piao Z. 2018. Identification and mapping of the clubroot resistance gene CRd in Chinese cabbage(Brassica rapa L. ssp. pekinensis). Frontiers in Plant Science, 9:653.
doi: 10.3389/fpls.2018.00653 URL |
| [11] |
Piao Z, Deng Y, Choi S, Park Y, Lim Y. 2004. SCAR and CAPS mapping of CRb,a gene conferring resistance to Plasmodiophora brassicae in Chinese cabbage(Brassica rapa ssp. pekinensis). Theoretical Applied Genetics, 108:1458-1465.
doi: 10.1007/s00122-003-1577-5 URL |
| [12] |
Sakamoto K, Saito A, Hayashida N, Taguchi G, Matsumoto E. 2008. Mapping of isolate-specific QTLs for clubroot resistance in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Theoretical Applied Genetics, 117:759-767.
doi: 10.1007/s00122-008-0817-0 URL |
| [13] |
Semagn K, Babu R, Hearne S, Olsen M. 2014. Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR(KASP):Overview of the technology and its application in crop improvement. Molecular Breeding, 33(1):1-14.
doi: 10.1007/s11032-013-9917-x URL |
| [14] | Sun Baoya, Shen Xiangqun, Guo Haifeng, Zhou Yonghong. 2005. Research progress in clubroot of crucifers and resistance breeding. China Vegetables, (4):34-37. (in Chinese) |
| 孙保亚, 沈向群, 郭海峰, 周永红. 2005. 十字花科植物根肿病及抗病育种研究进展. 中国蔬菜, (4):34-37. | |
| [15] |
Suwabe K, Suzuki G, Nunome T, Hatakeyama K, Mukai Y, Fukuoka H, Matsumoto S. 2012. Microstructure of a Brassica rapa genome segment homoeologous to the resistance gene cluster on Arabidopsis chromosome 4. Breeding Science, 62:170-177.
doi: 10.1270/jsbbs.62.170 URL pmid: 23136528 |
| [16] |
Suwabe K, Tsukazaki H, Iketani H, Hatakeyama K, Fujimura M, Nunome T, Fukuoka H, Matsumoto S, Hirai M. 2003. Identification of two loci for resistance to clubroot(Plasmodiophora brassicae Woron.)in Brassica rapa L. Theoretical Applied Genetics, 107:997-1002.
doi: 10.1007/s00122-003-1309-x URL |
| [17] |
Suwabe K, Tsukazaki H, Iketani H, Hatakeyama K, Kondo M, Fujimura M, Nunome T, Fukuoka H, Hirai M, Matsumoto S. 2006. Simple sequence repeat-based comparative genomics between Brassica rapa and Arabidopsis thaliana:the genetic origin of clubroot resistance. Genetics, 173:309-319.
pmid: 16723420 |
| [18] |
Takagi H, Abe A, Yoshida K, Kosugi S, Natsume S. 2013. QTLseq:rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. The Plant Journal, 74:174-183.
doi: 10.1111/tpj.2013.74.issue-1 URL |
| [19] |
Wang X Brassica rapa Genome Sequencing Project Consortium. 2011. The genome of the mesopolyploid crop species Brassica rapa. Nature Genetics, 43:1035-1039.
doi: 10.1038/ng.919 URL |
| [20] |
Wei X C, Xu W, Yuan Y X, Yao Q J, Zhao Y Y, Wang Z Y, Jiang W S, Zhang X W. 2016. Genome-wide investigation of microRNAs and their targets in Brassica rapa ssp. pekinensis root with Plasmodiophora brassicae infection. Horticultural Plant Journal, 2(4):209-216.
doi: 10.1016/j.hpj.2016.11.004 URL |
| [21] | Yang Limei, Fang Zhiyuan, Zhang Yangyong, Zhuang Mu, Lü Honghao, Wang Yong, Ji Jialei, Liu Yumei, Li Zhansheng, Han Fengqing. 2020. Recent advances of disease and stress resistance breeding of cabbage in China. Acta Horticulturae Sinica, 47(9):1678-1688. (in Chinese) |
| 杨丽梅, 方智远, 张扬勇, 庄木, 吕红豪, 王勇, 季家磊, 刘玉梅, 李占省, 韩风庆. 2020. 中国结球甘蓝抗病抗逆遗传育种近年研究进展. 园艺学报, 47(9):1678-1688. | |
| [22] | Yang Shaungjuan, Yuan Yuxiang, Wei Xiaochun, Wang Zhiyong, Zhao Yanyan, Yao Qiuju, Zhang Xiaowei. 2018. Optimization and establishment of KASP reaction system for Chinese cabbage(Brassica rapa L. ssp. pekinensis). Acta Horticulturae Sinica, 45(12):2442-2452. (in Chinese) |
| 杨双娟, 原玉香, 魏小春, 王志勇, 赵艳艳, 姚秋菊, 张晓伟. 2018. 大白菜 KASP 反应体系的优化与建立. 园艺学报, 45(12):2442-2452. | |
| [23] |
Yu F, Zhang X, Peng G, Falk K C, Strelkov S E, Gossen B D. 2017. Genotyping by sequencing reveals three QTL for clubroot resistance to six pathotypes of Plasmodiophora brassicae in Brassica rapa. Scientific Reports, 7(1):4516.
doi: 10.1038/s41598-017-04903-2 URL |
| [24] | Yuan Yuxiang, Zhao Yanyan, Wei Xiaochun, Yao Qiuju, Jiang Wusheng, Wang Zhiyong, Li Yang, Xu Qian, Yang Shuangjuan, Zhang Xiaowei. 2017. Pathotype identification of Plasmodiophora brassicae Woron. collected from Chinese cabbage in Henan Province. Journal of Henan Agricultural Sciences, 46(7):71-76. (in Chinese) |
| 原玉香, 赵艳艳, 魏小春, 姚秋菊, 蒋武生, 王志勇, 李扬, 许茜, 杨双娟, 张晓伟. 2017. 河南省大白菜根肿病菌生理小种鉴定. 河南农业科学, 46(7):71-76. | |
| [25] | Zhang Hui, Zhang Shujiang, Li Fei, Zhang Shifan, Li Guoliang, Ma Xiaochao, Liu Xitong, Sun Rifei. 2020. Research progress on clubroot disease resistance breeding of Brassica rapa. Acta Horticulturae Sinica, 47(9):1648-1662. (in Chinese) |
| 张慧, 张淑江, 李菲, 章时蕃, 李国亮, 马小超, 刘希童, 孙日飞. 2020. 大白菜抗根肿病育种研究进展. 园艺学报, 47(9):1648-1662. | |
| [26] | Zhang Jing, Wu Yue, Feng Hui, Ge Wenjie, Liu Xuyao, Lü Mingcan, Wang Yilian, Ji Ruiqin. 2019. Obtaining and identification of single-spore physiological races of Plasmodiophora brassicae in Williams system. Acta Horticulturae Sinica, 46(12):2415-2422. (in Chinese) |
| 张晶, 武月, 冯辉, 葛文杰, 刘旭垚, 吕明灿, 王艺琏, 冀瑞琴. 2019. 根肿菌Williams系统中各单孢生理小种的获得与鉴定. 园艺学报, 46(12):2415-2422. |
| [1] | 韩书辉, 韩彩锋, 韩书荣, 韩彩梅, 韩 旭. 秋大白菜新品种‘胶研秋宝’[J]. 园艺学报, 2022, 49(S2): 83-84. |
| [2] | 汪维红, 张凤兰, 余阳俊, 张德双, 赵岫云, 于拴仓, 苏同兵, 李佩荣, 辛晓云. 秋大白菜新品种‘京秋1518’[J]. 园艺学报, 2022, 49(S2): 85-86. |
| [3] | 余阳俊, 汪维红, 苏同兵, 张凤兰, 张德双, 赵岫云, 于拴仓, 李佩荣, 辛晓云, 王 姣. 抗根肿病耐抽薹大白菜新品种‘京春CR3’[J]. 园艺学报, 2022, 49(S2): 87-88. |
| [4] | 王丽丽, 王 鑫, 吴海东, 温 蔷, 杨晓飞. 抗根肿病大白菜新品种‘辽白28’[J]. 园艺学报, 2022, 49(S2): 89-90. |
| [5] | 余阳俊, 苏同兵, 张凤兰, 张德双, 赵岫云, 于拴仓, 汪维红, 李佩荣, 辛晓云, 王 姣, 武长见. 紫色苗用型大白菜新品种‘京研紫快菜’[J]. 园艺学报, 2022, 49(S2): 91-92. |
| [6] | 徐立功, 韩太利, 孙继峰, 杨晓东, 谭金霞. 苗用大白菜新品种‘锦绿2号’[J]. 园艺学报, 2022, 49(S1): 67-68. |
| [7] | 王钰, 张雪, 张学颖, 张思雨, 闻婷婷, 王迎君, 甘彩霞, 庞文星. 抗毒素Camalexin对大白菜抗根肿病的作用研究[J]. 园艺学报, 2022, 49(8): 1689-1698. |
| [8] | 张鲁刚, 卢倩倩, 何琼, 薛一花, 马晓敏, 马帅, 聂姗姗, 杨文静. 紫橙色大白菜新种质的创制[J]. 园艺学报, 2022, 49(7): 1582-1588. |
| [9] | 芮婷婷, 高青云, 李晓菁, 石延霞, 谢学文, 李磊, 张红杰, 徐文娟, 柴阿丽, 李宝聚. 芸薹根肿菌单孢亚甲基蓝琼脂糖分离法及其对生理小种的鉴定[J]. 园艺学报, 2022, 49(6): 1290-1300. |
| [10] | 乔军, 刘婧, 李素文, 王利英. 基于极端混合池全基因组重测序的茄子萼下果色基因预测[J]. 园艺学报, 2022, 49(3): 613-621. |
| [11] | 王光鹏, 刘同坤, 徐新凤, 李竹帛, 高瞻远, 侯喜林. 大白菜LEA家族基因的鉴定及其部分成员在低温胁迫下的表达分析[J]. 园艺学报, 2022, 49(2): 304-318. |
| [12] | 盛云燕, 杨丽敏, 戴冬洋, 张佳欣, 王岭, 王迪, 才羿, 田丽美. 甜瓜果柄离区细胞学观察及成熟脱落基因AL3的初步定位[J]. 园艺学报, 2022, 49(2): 341-351. |
| [13] | 王荣花, 王树彬, 刘栓桃, 李巧云, 张志刚, 王立华, 赵智中. 大白菜花茎蜡粉近等基因系转录组分析[J]. 园艺学报, 2022, 49(1): 62-72. |
| [14] | 徐立功, 韩太利, 孙继峰, 杨晓东, 谭金霞, 宋银行, 李 萌, 周陆红, 孙莎莎. 春大白菜新品种‘潍春22号’[J]. 园艺学报, 2021, 48(S2): 2825-2826. |
| [15] | 汪维红, 张凤兰, 余阳俊, 张德双, 赵岫云, 于拴仓, 苏同兵, 李佩荣, 辛晓云. 秋大白菜新品种‘京秋5号’[J]. 园艺学报, 2021, 48(S2): 2827-2828. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司