
园艺学报 ›› 2021, Vol. 48 ›› Issue (6): 1053-1066.doi: 10.16420/j.issn.0513-353x.2020-0653
• 研究论文 • 下一篇
收稿日期:2021-02-07
修回日期:2021-05-11
出版日期:2021-06-25
发布日期:2021-07-07
通讯作者:
程云清
E-mail:Chengyunqing1977@163.com
基金资助:
LIU Jianfeng, SUN Ying, WEI Heng, HE Hongli, ZHANG Xingzheng, CHENG Yunqing( )
)
Received:2021-02-07
Revised:2021-05-11
Online:2021-06-25
Published:2021-07-07
Contact:
CHENG Yunqing
E-mail:Chengyunqing1977@163.com
摘要:
在榛子胚珠不同发育时期筛选差异表达的circRNA,进一步鉴定其差异表达的靶mRNA,以期明确circRNA在胚珠中的表达特征及其对胚珠发育的调控作用。在榛子胚珠形成期(Ov1)、早期生长期(Ov2)、快速生长期(Ov3)和成熟期(Ov4)进行胚珠样品采集和Illumina高通量测序,结合已有的mRNA测序分析结果,鉴定参与胚珠发育的差异表达circRNA(DEcircRNA)及其靶差异表达mRNA(DEmRNA)。共鉴定到7 122条circRNA,59.54%的circRNA来自外显子区域。在成对比较Ov1-vs-Ov2、Ov1-vs-Ov3、Ov1-vs-Ov4、Ov2-vs-Ov3、Ov2-vs-Ov4和Ov3-vs-Ov4中,分别鉴定到123、162、187、85、105和21条DEcircRNA;DEcircRNA的表达有明显的发育时期特异性;有199条DEcircRNA的靶mRNA也发生差异表达,两者表达变化高度同步。GO(Gene Ontology)分类结果表明DEcircRNA参与胚珠发育过程中代谢的调控。随机选择了4个差异表达circRNA及靶mRNA,用qRT-PCR分析其在4个胚珠发育阶段表达量的变化,证实qRT-PCR与Illumina高通量测序结果相一致。
中图分类号:
刘剑锋, 孙莹, 魏珩, 贺红利, 张兴政, 程云清. 榛子胚珠不同发育阶段circRNA的分析与鉴定[J]. 园艺学报, 2021, 48(6): 1053-1066.
LIU Jianfeng, SUN Ying, WEI Heng, HE Hongli, ZHANG Xingzheng, CHENG Yunqing. Analysis and Identification of circRNAs of Hazel Ovule at Different Developmental Stages[J]. Acta Horticulturae Sinica, 2021, 48(6): 1053-1066.
| RNA | 基因名称 Gene ID | 引物序列(5′-3′) Primer sequence | 产物长度/bp Amplicon size |
|---|---|---|---|
| DEcircRNA | 00262:4702|5542 | F:GTTCTGTTCCCTGCCTCCAAT;R:CCATTTTCCCAACATTCCTTAGT | 129 |
| 03593:4109|4340 | F:AAACAAGAGGCCGAAACTAG;R:ATCCATTGTCTGCAAGTGTT | 115 | |
| 05162:7223|7649 | F:CAGTTCAGGGTAGTTGTCG;R:GTTTGCTAGGCTTAGATACTTC | 103 | |
| 05531:13219|13378 | F:TGTGCCCTTCATGTTCTCCTT;R:CACGAGGAGGATTGGAGGAAG | 133 | |
| DEmRNA | g3238 | F:CGGTAATAAGAGCCCAGGTGC;R:CACCTGTGCCGTGAATTTGTC | 142 |
| g18070 | F:CGCTCTTGCTCAGGCTTGTC;R:GAGAAGGCATGGATACCCGTC | 87 | |
| g21729 | F:CTGCAAAGAACTCCCTGTGGTAA;R:TTGGCAGGGCTCAGGCATT | 129 | |
| g22409 | F:GCGGTATCAAGAACCAAAGGG;R:GAGCGTCAAACCAGCAAGTG | 110 | |
| 内参基因 Reference gene | Beta actin | F:CCCTCACAATTTCACGCTCG;R:ATGAGGGTTATGCCCTCCCA | 135 |
表1 DEcircRNA与其靶DEmRNA实时荧光定量分析引物序列
Table 1 Primer sequences of DEcircRNAs and their target DEmRNAs in qRT-PCR analysis
| RNA | 基因名称 Gene ID | 引物序列(5′-3′) Primer sequence | 产物长度/bp Amplicon size |
|---|---|---|---|
| DEcircRNA | 00262:4702|5542 | F:GTTCTGTTCCCTGCCTCCAAT;R:CCATTTTCCCAACATTCCTTAGT | 129 |
| 03593:4109|4340 | F:AAACAAGAGGCCGAAACTAG;R:ATCCATTGTCTGCAAGTGTT | 115 | |
| 05162:7223|7649 | F:CAGTTCAGGGTAGTTGTCG;R:GTTTGCTAGGCTTAGATACTTC | 103 | |
| 05531:13219|13378 | F:TGTGCCCTTCATGTTCTCCTT;R:CACGAGGAGGATTGGAGGAAG | 133 | |
| DEmRNA | g3238 | F:CGGTAATAAGAGCCCAGGTGC;R:CACCTGTGCCGTGAATTTGTC | 142 |
| g18070 | F:CGCTCTTGCTCAGGCTTGTC;R:GAGAAGGCATGGATACCCGTC | 87 | |
| g21729 | F:CTGCAAAGAACTCCCTGTGGTAA;R:TTGGCAGGGCTCAGGCATT | 129 | |
| g22409 | F:GCGGTATCAAGAACCAAAGGG;R:GAGCGTCAAACCAGCAAGTG | 110 | |
| 内参基因 Reference gene | Beta actin | F:CCCTCACAATTTCACGCTCG;R:ATGAGGGTTATGCCCTCCCA | 135 |
| 注释数据库Annotation database | 注释数量Annotated number | 基因长度Gene length | |
|---|---|---|---|
| (300 ~ 999 bp) | (> = 1 000 bp) | ||
| COG | 1 440 | 103 | 1 335 |
| GO | 2 398 | 263 | 2 130 |
| KEGG | 1 741 | 188 | 1 548 |
| KOG | 2 377 | 241 | 2 133 |
| Pfam | 2 859 | 243 | 2 616 |
| Swissprot | 2 793 | 280 | 2 509 |
| eggNOG | 3 440 | 367 | 3 068 |
| NR | 3 509 | 391 | 3 113 |
| 总数 All annotated | 3 511 | 391 | 3 115 |
表2 circRNA的注释
Table 2 Annotation of identified circRNAs
| 注释数据库Annotation database | 注释数量Annotated number | 基因长度Gene length | |
|---|---|---|---|
| (300 ~ 999 bp) | (> = 1 000 bp) | ||
| COG | 1 440 | 103 | 1 335 |
| GO | 2 398 | 263 | 2 130 |
| KEGG | 1 741 | 188 | 1 548 |
| KOG | 2 377 | 241 | 2 133 |
| Pfam | 2 859 | 243 | 2 616 |
| Swissprot | 2 793 | 280 | 2 509 |
| eggNOG | 3 440 | 367 | 3 068 |
| NR | 3 509 | 391 | 3 113 |
| 总数 All annotated | 3 511 | 391 | 3 115 |

图3 不同发育阶段差异表达circRNA的系统聚类分析 A、B和C为3次生物学重复。
Fig. 3 Hierarchical clustering analysis of differentially expressed circRNAs at four developmental stages. A,B and C represent three biological repetitions.
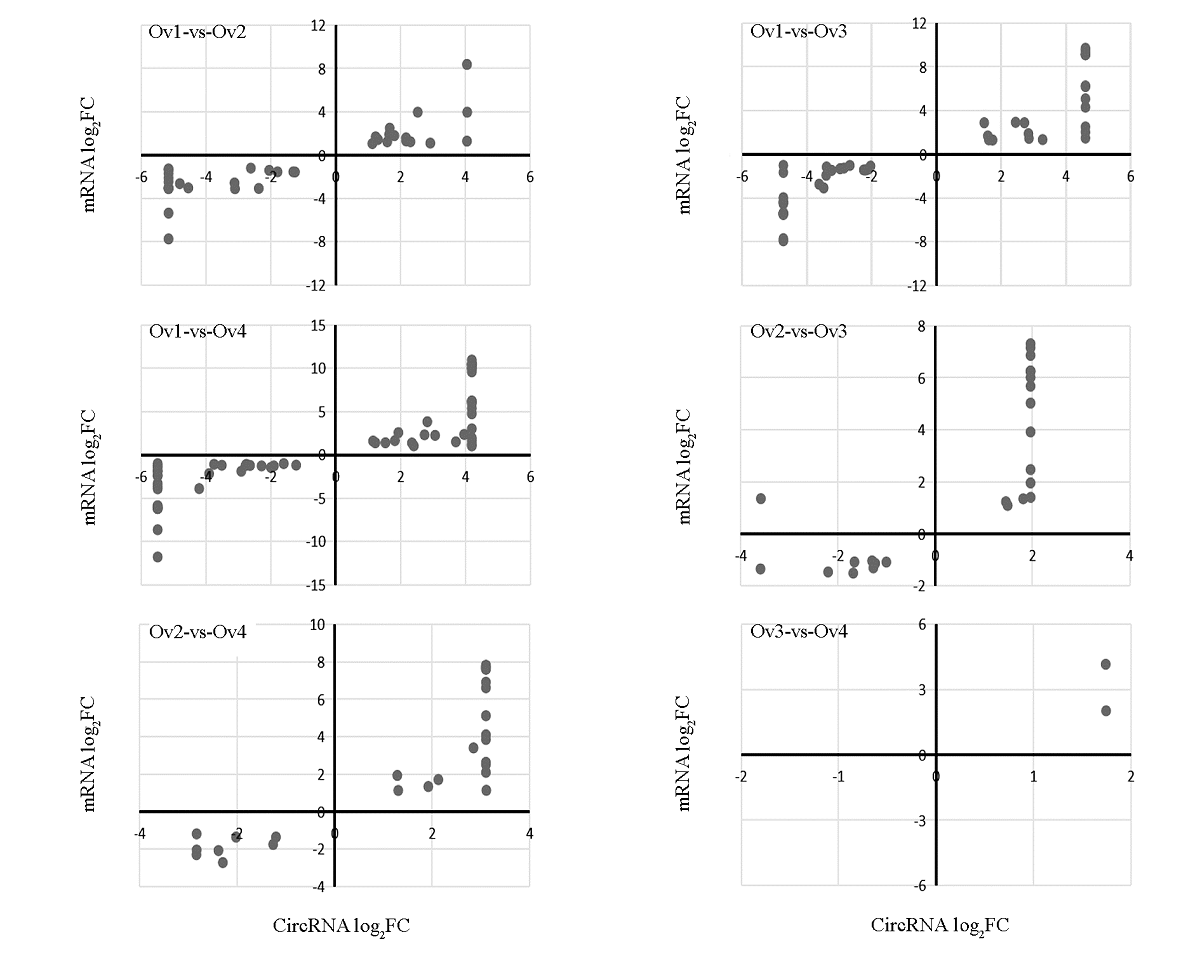
图4 DEcircRNA与其靶DEmRNA的关联分析 在成对比较中,FC表示后一个发育阶段中circRNA表达量(B)/前一个发育阶段中circRNA表达量(A),如果B为0,log2FC = -∞,如果A为0,log2FC = ∞,为将每对DEcircRNA-DEmRNA在图中直观地表示出来,当log 2FC = -∞或∞时,将log2FC的值分别记为该成对比较中的最小值与最大值。
Fig. 4 Correlation analysis of DEcircRNA and their target DEmRNA In paired comparison,FC represents circRNA expression in latter development stage(B)/ circRNA expression in former development stage(A). If B is 0,log2FC = -∞;if A is 0,log2FC = ∞. In order to express each pair of DEcircRNA-DEmRNA in the graph intuitively,when log 2FC = -∞ or ∞,the value of log2FC is recorded as the minimum and maximum value in the paired comparison respectively.

图6 DEcircRNA与其靶DEmRNA实时荧光定量分析 同一折线中不同小写字母表示差异显著,P < 0.05。
Fig. 6 Analysis of DEcircRNAs and their target DEmRNAs using qRT-PCR Different lowercase letters in the same broken line indicates significant difference at level of P < 0.05.
| 成对比较 Paired comparison | 差异表达circRNA DEcircRNA | log2FC (circRNA) | 差异表达mRNA DEmRNA | log2FC (mRNA) | 注释 Annotation |
|---|---|---|---|---|---|
| Ov1-vs-Ov4 | 19296:1590|1950 | -∞ | Corylus_avellana_newGene_15264 | -11.80 | NPR4 |
| Ov1-vs-Ov2 | 19296:1590|1950 | -∞ | Corylus_avellana_newGene_15264 | -7.74 | NPR4 |
| Ov1-vs-Ov3 | 19296:1590|1950 | -∞ | Corylus_avellana_newGene_15264 | -7.73 | NPR4 |
| Ov1-vs-Ov4 | 00262:4702|5542 | -∞ | g3238 | -6.19 | NPP |
| Ov1-vs-Ov4 | 00262:4702|5735 | -∞ | g3238 | -6.19 | NPP |
| Ov1-vs-Ov4 | 00262:5280|5910 | -∞ | g3238 | -6.19 | NPP |
| Ov1-vs-Ov3 | 01486:5071|5547 | -∞ | g10760 | -4.39 | ABCG9 |
| Ov1-vs-Ov3 | 09164:4340|5637 | -∞ | g27362 | -4.32 | SULTR3;1 |
| Ov1-vs-Ov3 | 05982:1271|3603 | -∞ | Corylus_avellana_newGene_58963 | -3.97 | SCPL18 |
| Ov1-vs-Ov4 | 09164:4340|5637 | -∞ | g27362 | -3.93 | SULTR3;1 |
| Ov1-vs-Ov4 | 05162:7223|7649 | -4.21 | g21729 | -3.90 | SGR |
| Ov1-vs-Ov4 | 05982:1271|3603 | -∞ | Corylus_avellana_newGene_58963 | -3.64 | SCPL18 |
| Ov1-vs-Ov3 | 05162:7223|7649 | -3.48 | g21729 | -3.08 | SGR |
| Ov1-vs-Ov2 | 00262:4702|5542 | -∞ | g3238 | -3.03 | NPP |
| Ov1-vs-Ov2 | 00262:4702|5735 | -4.55 | g3238 | -3.03 | NPP |
| Ov2-vs-Ov4 | 05312:1137|1462 | ∞ | Corylus_avellana_newGene_56641 | 6.62 | NHX4 |
| Ov2-vs-Ov3 | 14244:2582|2731 | ∞ | g31169 | 6.87 | OLE1 |
| Ov2-vs-Ov4 | 01490:2219|2537 | ∞ | g10779 | 6.92 | COR2 |
| Ov2-vs-Ov3 | 03593:4109|4340 | ∞ | g18070 | 7.31 | TIP3-2 |
| Ov2-vs-Ov4 | 03593:4109|4340 | ∞ | g18070 | 7.63 | TIP3-2 |
| 成对比较 Paired comparison | 差异表达circRNA DEcircRNA | log2FC (circRNA) | 差异表达mRNA DEmRNA | log2FC (mRNA) | 注释 Annotation |
| Ov2-vs-Ov4 | 03974:6066|6673 | ∞ | g19072 | 7.80 | SBP |
| Ov2-vs-Ov4 | 14244:2582|2731 | ∞ | g31169 | 7.82 | OLE1 |
| Ov1-vs-Ov2 | 04715:14790|15271 | ∞ | g20792 | 8.37 | WAT1 |
| Ov1-vs-Ov3 | 03593:4109|4340 | ∞ | g18070 | 9.34 | TIP3-2 |
| Ov1-vs-Ov3 | 14244:2582|2731 | ∞ | g31169 | 9.55 | OLE1 |
| Ov1-vs-Ov4 | 03593:4109|4340 | ∞ | g18070 | 9.59 | TIP3-2 |
| Ov1-vs-Ov3 | 05531:13219|13378 | ∞ | g22409 | 9.69 | OLE5 |
| Ov1-vs-Ov4 | 05531:13219|13378 | ∞ | g22409 | 9.99 | OLE5 |
| Ov1-vs-Ov4 | 05531:13219|13433 | ∞ | g22409 | 9.99 | OLE5 |
| Ov1-vs-Ov4 | 14244:2582|2731 | ∞ | g31169 | 10.35 | OLE1 |
表3 表达量变化最大的30对DEcircRNA及其靶DEmRNA
Table 3 Thirty pairs of DEcircRNAs with the largest expression fold change and their target DEmRNAs
| 成对比较 Paired comparison | 差异表达circRNA DEcircRNA | log2FC (circRNA) | 差异表达mRNA DEmRNA | log2FC (mRNA) | 注释 Annotation |
|---|---|---|---|---|---|
| Ov1-vs-Ov4 | 19296:1590|1950 | -∞ | Corylus_avellana_newGene_15264 | -11.80 | NPR4 |
| Ov1-vs-Ov2 | 19296:1590|1950 | -∞ | Corylus_avellana_newGene_15264 | -7.74 | NPR4 |
| Ov1-vs-Ov3 | 19296:1590|1950 | -∞ | Corylus_avellana_newGene_15264 | -7.73 | NPR4 |
| Ov1-vs-Ov4 | 00262:4702|5542 | -∞ | g3238 | -6.19 | NPP |
| Ov1-vs-Ov4 | 00262:4702|5735 | -∞ | g3238 | -6.19 | NPP |
| Ov1-vs-Ov4 | 00262:5280|5910 | -∞ | g3238 | -6.19 | NPP |
| Ov1-vs-Ov3 | 01486:5071|5547 | -∞ | g10760 | -4.39 | ABCG9 |
| Ov1-vs-Ov3 | 09164:4340|5637 | -∞ | g27362 | -4.32 | SULTR3;1 |
| Ov1-vs-Ov3 | 05982:1271|3603 | -∞ | Corylus_avellana_newGene_58963 | -3.97 | SCPL18 |
| Ov1-vs-Ov4 | 09164:4340|5637 | -∞ | g27362 | -3.93 | SULTR3;1 |
| Ov1-vs-Ov4 | 05162:7223|7649 | -4.21 | g21729 | -3.90 | SGR |
| Ov1-vs-Ov4 | 05982:1271|3603 | -∞ | Corylus_avellana_newGene_58963 | -3.64 | SCPL18 |
| Ov1-vs-Ov3 | 05162:7223|7649 | -3.48 | g21729 | -3.08 | SGR |
| Ov1-vs-Ov2 | 00262:4702|5542 | -∞ | g3238 | -3.03 | NPP |
| Ov1-vs-Ov2 | 00262:4702|5735 | -4.55 | g3238 | -3.03 | NPP |
| Ov2-vs-Ov4 | 05312:1137|1462 | ∞ | Corylus_avellana_newGene_56641 | 6.62 | NHX4 |
| Ov2-vs-Ov3 | 14244:2582|2731 | ∞ | g31169 | 6.87 | OLE1 |
| Ov2-vs-Ov4 | 01490:2219|2537 | ∞ | g10779 | 6.92 | COR2 |
| Ov2-vs-Ov3 | 03593:4109|4340 | ∞ | g18070 | 7.31 | TIP3-2 |
| Ov2-vs-Ov4 | 03593:4109|4340 | ∞ | g18070 | 7.63 | TIP3-2 |
| 成对比较 Paired comparison | 差异表达circRNA DEcircRNA | log2FC (circRNA) | 差异表达mRNA DEmRNA | log2FC (mRNA) | 注释 Annotation |
| Ov2-vs-Ov4 | 03974:6066|6673 | ∞ | g19072 | 7.80 | SBP |
| Ov2-vs-Ov4 | 14244:2582|2731 | ∞ | g31169 | 7.82 | OLE1 |
| Ov1-vs-Ov2 | 04715:14790|15271 | ∞ | g20792 | 8.37 | WAT1 |
| Ov1-vs-Ov3 | 03593:4109|4340 | ∞ | g18070 | 9.34 | TIP3-2 |
| Ov1-vs-Ov3 | 14244:2582|2731 | ∞ | g31169 | 9.55 | OLE1 |
| Ov1-vs-Ov4 | 03593:4109|4340 | ∞ | g18070 | 9.59 | TIP3-2 |
| Ov1-vs-Ov3 | 05531:13219|13378 | ∞ | g22409 | 9.69 | OLE5 |
| Ov1-vs-Ov4 | 05531:13219|13378 | ∞ | g22409 | 9.99 | OLE5 |
| Ov1-vs-Ov4 | 05531:13219|13433 | ∞ | g22409 | 9.99 | OLE5 |
| Ov1-vs-Ov4 | 14244:2582|2731 | ∞ | g31169 | 10.35 | OLE1 |
| [1] |
Ambros V R. 2004. The functions of animal microRNAs. Nature, 431:350-355.
doi: 10.1038/nature02871 URL |
| [2] | An Lin-jun, Luan Jia-yu, Ren Li, Li Hui-yu, Xia De-an. 2019. Bioinformatics and expression characteristics analysis of BpTCP8 in Betula platyphylla Suk. Journal of Nanjing Forestry Univeristy(Natural Sciences Edition), 43:67-73. (in Chinese) |
| 安琳君, 栾嘉豫, 任丽, 李慧玉, 夏德安. 2019. 白桦 BpTCP8基因生物信息学及表达特性分析. 南京林业大学学报(自然科学版), 43:67-73. | |
| [3] |
Apweiler R, Bairoch A M, Wu C H, Barker W C, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M. 2004. UniProt:the universal protein knowledgebase. Nucleic Acids Research, 32:115-119.
doi: 10.1093/nar/gkh151 URL |
| [4] |
Ashburner M, Ball C A, Blake J A, Botstein D, Butler H, Cherry J M, Davis A P, Dolinski K, Dwight S S, Eppig J T, Harris M A, Hill D P, Issel-Tarver L, Kasarskis A, Lewis S, Matese J C, Richardson J E, Ringwald M, Rubin G M, Sherlock G. 2000. Gene ontology:tool for the unification of biology. Nature Genetics, 25:25-29.
doi: 10.1038/75556 URL |
| [5] |
Ashwalfluss R, Meyer M, Pamudurti N R, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. 2014. circRNA biogenesis competes with pre-mRNA splicing. Molecular Cell, 56:55-66.
doi: 10.1016/j.molcel.2014.08.019 URL |
| [6] |
Bartel D P. 2004. microRNAs:genomics,biogenesis,mechanism,and function. Cell, 116:281-297.
pmid: 14744438 |
| [7] |
Bartel D P. 2009. microRNAs:target recognition and regulatory functions. Cell, 136:215-233.
doi: 10.1016/j.cell.2009.01.002 pmid: 19167326 |
| [8] |
Bartel D P. 2018. Metazoan microRNAs. Cell, 173:20-51.
doi: S0092-8674(18)30286-1 pmid: 29570994 |
| [9] |
Cao H P, Zhang L, Tan X F, Long H X, Shockey J M. 2014. Identification,classification and differential expression of oleosin genes in tung tree (Vernicia fordii). PLoS ONE, 9:e88409.
doi: 10.1371/journal.pone.0088409 URL |
| [10] |
Cao M J, Wang Z, Wirtz M, Hell R, Oliver D J, Xiang C B. 2013. SULTR3;1 is a chloroplast-localized sulfate transporter in Arabidopsis thaliana. Plant Journal, 73:607-616.
doi: 10.1111/tpj.2013.73.issue-4 URL |
| [11] | Cheng Yun-qing, Zhang Li-na, Zhao Yong-bin, Liu Jian-feng. 2018. Analysis of SSR markers information and primer selection from transcriptome sequence of hybrid hazelnut Corylus heterophylla × C. avellana. Acta Horticulturae Sinica, 45 (1):139-148. (in Chinese) |
| 程云清, 张丽娜, 赵永斌, 刘剑锋. 2018. 平欧杂交榛转录组中SSR信息分析和引物筛选. 园艺学报, 45 (1):139-148. | |
| [12] | Deng Yang-yang, Li Jian-qi, Wu Song-feng, Zhu Yun-ping, Chen Yao-wen, He Fu-chu. 2006. Integrated nr database in protein annotation system and its localization. Computer Engineering, 32:71-74. (in Chinese) |
| 邓泱泱, 荔建琦, 吴松锋, 朱云平, 陈耀文, 贺福初. 2006. nr数据库分析及其本地化. 计算机工程, 32:71-73. | |
| [13] |
Eddy S R. 1998. Profile hidden Markov models. Bioinformatics, 14:755-763.
pmid: 9918945 |
| [14] | Gao Zhen, Luo Meng, Wang Lei, Song Shiren, Zhao Liping, Xu Wenping, Zhang Caixi, Wang Shiping, Ma Chao. 2019. The research advance of plant circular RNA. Acta Horticulturae Sinica, 46 (1):171-181. |
| 高振, 骆萌, 王磊, 宋士任, 赵丽萍, 许文平, 张才喜, 王世平, 马超. 2019. 植物环状RNA研究进展. 园艺学报, 46 (1):171-181. | |
| [15] |
Hansen T B, Jensen T I, Clausen B H, Bramsen J B, Finsen B, Damgaard C K, Kjems J. 2013. Natural RNA circles function as efficient microRNA sponges. Nature, 495:384-388.
doi: 10.1038/nature11993 URL |
| [16] | Hu Xiao-yi, Tan Xiao-feng, Tian Xiao-ming, Liu Qiao, Luo Qian, Chen Hong-peng, Hu Fang-min. 2008. Identification and analysis of an aquaporin (CoPIP1-1)in the seeds of Camellia oleifer. Scientia Silvae Sinicae, 44:48-56. (in Chinese) |
| 胡孝义, 谭晓风, 田晓明, 刘巧, 罗茜, 陈鸿鹏, 胡芳名. 2008. 油茶种子水通道蛋白CoPIP1-1的鉴定与分析. 林业科学, 44:51-59. | |
| [17] |
Jeck W R, Sharpless N E. 2014. Detecting and characterizing circular RNAs. Nature Biotechnology, 32:453-461.
doi: 10.1038/nbt.2890 URL |
| [18] | Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. 2004. The KEGG resource for deciphering the genome. Nucleic Acids Research, 32:277-280. |
| [19] | Koonin E V, Fedorova N D, Jackson J D, Jacobs A R, Krylov D M, Makarova K S, Mazumder R, Mekhedov S L, Nikolskaya A N, Rao B S. 2004. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biology, 5:1-28. |
| [20] |
Kristensen L S, Andersen M S, Stagsted L V W, Ebbesen K K, Hansen T B, Kjems J. 2019. The biogenesis,biology and characterization of circular RNAs. Nature Reviews Genetics, 20:675-691.
doi: 10.1038/s41576-019-0158-7 |
| [21] |
Le Hir R, Sorin C, Chakraborti D, Moritz T, Schaller H, Tellier F, Robert S, Morin H, Bako L, Bellini C. 2013. ABCG9,ABCG11 and ABCG14 ABC transporters are required for vascular development in Arabidopsis. Plant Journal, 76:811-824.
doi: 10.1111/tpj.12334 URL |
| [22] |
Li B, Ruotti V, Stewart R M, Thomson J A, Dewey C N. 2010. RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics, 26:493-500.
doi: 10.1093/bioinformatics/btp692 URL |
| [23] | Li J Q, Yang J, Zhou P, Le Y P, Zhou C W, Wang S M, Xu D Z, Lin H K, Gong Z H. 2015a. Circular RNAs in cancer:novel insights into origins,properties,functions and implications. American Journal of Cancer Research, 5:472-480. |
| [24] |
Li Y B, Fan C C, Xing Y Z, Jiang Y H, Luo L J, Sun L, Shao D, Xu C J, Li X H, Xiao J H. 2011. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nature Genetics, 43:1266-1269.
doi: 10.1038/ng.977 URL |
| [25] |
Li Z Y, Huang C, Bao C, Chen L, Lin M, Wang X L, Zhong G L, Yu B, Hu W C, Dai L M. 2015b. Exon-intron circular RNAs regulate transcription in the nucleus. Nature Structural & Molecular Biology, 22:256-264.
doi: 10.1038/nsmb.2959 URL |
| [26] |
Liu J F, Cheng Y Q, Yan K, Liu Q, Wang Z W. 2012. The relationship between reproductive growth and blank fruit formation in Corylus heterophylla Fisch. Scientia Horticulturae, 136:128-134.
doi: 10.1016/j.scienta.2012.01.008 URL |
| [27] |
Liu J F, Luo Q Z, Zhang X Z, Zhang Q, Cheng Y Q. 2020. Identification of vital candidate microRNA/mRNA pairs regulating ovule development using high-throughput sequencing in hazel. BMC Developmental Biology, 20:13.
doi: 10.1186/s12861-020-00219-z URL |
| [28] |
Liu J F, Zhang H D, Cheng YQ, Kafkas S, Guney M. 2014. Pistillate flower development and pollen tube growth mode during the delayed fertilization stage in Corylus heterophylla Fisch. Plant Reproduction, 27:145-152.
doi: 10.1007/s00497-014-0248-9 URL |
| [29] |
Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods, 25:402-408.
pmid: 11846609 |
| [30] | Lü Dong-lin, Guo Yi-wen, Han Rui, Jiang Jing. 2018. Characterization of gene expression in anthocyanin synthesisand salt tolerance of Betula pendula 'Purple Rain'. Journal of Nanjing Forestry Univeristy(Natural Sciences Edition), 42:25-32. (in Chinese) |
| 吕东林, 林琳, 郭译文, 韩锐, 姜静. 2018. 紫雨桦耐盐性及花色苷合成相关基因的表达特性. 南京林业大学学报(自然科学版), 42:28-35. | |
| [31] |
Ma L, Li T, Hao C Y, Wang Y Q, Chen X H, Zhang X Y. 2016. TaGS5-3A,a grain size gene selected during wheat improvement for larger kernel and yield. Plant Biotechnology Journal, 14:1269-1280.
doi: 10.1111/pbi.2016.14.issue-5 URL |
| [32] |
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak S D, Gregersen L H, Munschauer M. 2013. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature, 495:333-338.
doi: 10.1038/nature11928 URL |
| [33] |
Nanjo Y, Oka H, Ikarashi N, Kaneko K, Kitajima A, Mitsui T, Munoz F, Rodriguezlopez M, Barojafernandez E, Pozuetaromero J. 2006. Rice plastidial N-glycosylated nucleotide pyrophosphatase/phosphodiesterase is transported from the ER-golgi to the chloroplast through the secretory pathway. Plant Cell, 18:2582-2592.
doi: 10.1105/tpc.105.039891 URL |
| [34] |
Philips A, Nowis K, Stelmaszczuk M, Podkowiński J, Handschuh L, Jackowiak P, Figlerowicz M. 2020. Arabidopsis thaliana cbp80, c2h2,and flk knockout mutants accumulate increased amounts of circular RNAs. Cells, 9:1937.
doi: 10.3390/cells9091937 URL |
| [35] |
Ranocha P, Dima O, Nagy R, Felten J, Corratgé-Faillie C, Novák O, Morreel K, Lacombe B, Martinez Y, Pfrunder S, Jin X, Renou J P, Thibaud J B, Ljung K, Fischer U, Martinoia E, Boerjan W, Goffner D. 2013. Arabidopsis WAT 1 is a vacuolar auxin transport facilitator required for auxin homoeostasis. Nature Communications, 4:2625.
doi: 10.1038/ncomms3625 URL |
| [36] | Shi Tian-pei, Wang Xin-yue, Hou Hao-bin, Zhao Zhi-da, Shang Ming-yu, Zhang Li. 2020. Analysis and identification of circRNAs of skeletal muscle at different stages of sheep embryos based on whole transcriptome sequencing. Scientia Agricultura Sinica, 53:642-657. (in Chinese) |
| 石田培, 王欣悦, 侯浩宾, 赵志达, 尚明玉, 张莉. 2020. 基于全转录组测序的绵羊胚胎不同发育阶段骨骼肌circRNA的分析与鉴定. 中国农业科学, 53:642-657. | |
| [37] |
Tan J J, Zhou Z J, Niu Y J, Sun X Y, Deng Z P. 2017. Identification and functional characterization of tomato circRNAs derived from genes involved in fruit pigment accumulation. Scientific Reports, 7:8594.
doi: 10.1038/s41598-017-08806-0 URL |
| [38] | Tatusov R L, Galperin M Y, Natale D A, Koonin E V. 2000. The COG database:a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Research, 28:33-36. |
| [39] |
Tay Y, Rinn J L, Pandolfi P P. 2014. The multilayered complexity of ceRNA crosstalk and competition. Nature, 505:344-352.
doi: 10.1038/nature12986 URL |
| [40] | Tian Xue-yao, Zhou Jie, Wang Bao-song, He Kai-yue, He Xu-dong. 2020. Cloning and expression pattern analysis of NAC genes in Salix. Journal of Nanjing Forestry Univeristy(Natural Sciences Edition), 44:123-128. (in Chinese) |
| 田雪瑶, 周洁, 王保松, 何开跃, 何旭东. 2020. 柳树 NAC基因的克隆与表达模式分析. 南京林业大学学报(自然科学版), 44:123-128. | |
| [41] | Wang H L, Xiao Y, Wu L, Ma D C. 2018. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-000911/miR-449a pathway in breast carcinogenesis. International Journal of Oncology, 52:743-754. |
| [42] |
Wang Y, Xiong Z Y, Li Q, Sun Y Y, Jin J, Chen H, Zou Y, Huang X G, Ding Y. 2019. Circular RNA profiling of the rice photo-thermosensitive genic male sterile line Wuxiang S reveals circRNA involved in the fertility transition. BMC Plant Biology, 19:340.
doi: 10.1186/s12870-019-1944-2 URL |
| [43] |
Wang Y X, Wang Q, Gao L P, Zhu B Z, Luo Y B, Deng Z P, Zuo J H. 2017a. Integrative analysis of circRNAs acting as ceRNAs involved in ethylene pathway in tomato. Physiologia Plantarum, 161:311-321.
doi: 10.1111/ppl.2017.161.issue-3 URL |
| [44] | Wang Z P, Liu Y F, Li D W, Li L, Zhang Q, Wang S B, Huang H W. 2017b. Identification of circular RNAs in kiwifruit and their species-specific response to bacterial canker pathogen invasion. Frontiers in Plant Science, 8:413. |
| [45] | Xiong H, Shekhar S, Tan P N, Kumar V. 2004. Exploiting a support-based upper bound of Pearson's correlation coefficient for efficiently identifying strongly correlated pairs//Proceedings of the tenth ACM SIGKDD international conference on knowledge discovery and data mining. Seattle:Association for Computing Machinery:334-343. |
| [46] |
Xu C J, Liu Y, Li Y B, Xu X D, Xu C G, Li X H, Xiao J H, Zhang Q F. 2015. Differential expression of GS5 regulates grain size in rice. Journal of Experimental Botany, 66:2611-2623.
doi: 10.1093/jxb/erv058 URL |
| [47] |
Yao J T, Zhao S H, Liu Q P, Lü M Q, Zhou D X, Liao Z J, Nan K J. 2017. Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathology-Research and Practice, 213:453-456.
doi: 10.1016/j.prp.2017.02.011 URL |
| [48] |
Zhang Y, Zhang X O, Chen T, Xiang J F, Yin Q F, Xing Y H, Zhu S S, Yang L, Chen L L. 2013. Circular intronic long noncoding RNAs. Molecular Cell, 51:792-806.
doi: 10.1016/j.molcel.2013.08.017 URL |
| [49] |
Zhao W, Cheng Y H, Zhang C, You Q B, Shen X J, Guo W, Jiao Y Q. 2017. Genome-wide identification and characterization of circular RNAs by high throughput sequencing in soybean. Scientific Reports, 7:5636.
doi: 10.1038/s41598-017-05922-9 URL |
| [50] |
Zhou R, Yu X Q, Ottosen C O, Zhao T M. 2020. High throughput sequencing of CircRNAs in tomato leaves responding to multiple stresses of drought and heat. Horticultural Plant Journal, 6 (1):34-38.
doi: 10.1016/j.hpj.2019.12.004 URL |
| [1] | 蒋 彧, 涂勋良, 何俊蓉. 国兰叶色突变体叶片差异表达基因分析[J]. 园艺学报, 2023, 50(2): 371-381. |
| [2] | 蔺海娇, 梁雨晨, 李玲, 马军, 张璐, 兰振颖, 苑泽宁. 薰衣草CBF途径相关耐寒基因挖掘与调控网络分析[J]. 园艺学报, 2023, 50(1): 131-144. |
| [3] | 赵雪艳, 王琪, 王莉, 王方圆, 王庆, 李艳. 基于比较转录组的延胡索组织差异性表达分析[J]. 园艺学报, 2023, 50(1): 177-187. |
| [4] | 周徐子鑫, 杨威, 毛美琴, 薛彦斌, 马均. 金边红苞凤梨叶色突变体色素鉴定及类胡萝卜素合成限速基因筛选[J]. 园艺学报, 2022, 49(5): 1081-1091. |
| [5] | 沈楠, 张荆城, 王成晨, 边银丙, 肖扬. 香菇子实体发育过程中的转录组研究[J]. 园艺学报, 2022, 49(4): 801-815. |
| [6] | 夏铭, 李经纬, 罗章瑞, 祖贵东, 王娅, 张万萍. 外源褪黑素影响萝卜生长及对链格孢菌抗性的机理研究[J]. 园艺学报, 2022, 49(3): 548-560. |
| [7] | 张瑞, 张夏燚, 赵婷, 王双成, 张仲兴, 刘博, 张德, 王延秀. 基于转录组分析垂丝海棠响应盐碱胁迫的分子机制[J]. 园艺学报, 2022, 49(2): 237-251. |
| [8] | 邓娇, 苏梦月, 刘雪莲, 欧克芳, 户正荣, 杨平仿. 基于转录组分析揭示双色花莲‘大洒锦’花色形成机理[J]. 园艺学报, 2022, 49(2): 365-377. |
| [9] | 乔军, 王利英, 刘婧, 李素文. 基于转录组测序的茄子萼下果色光敏相关基因表达分析[J]. 园艺学报, 2022, 49(11): 2347-2356. |
| [10] | 王荣花, 王树彬, 刘栓桃, 李巧云, 张志刚, 王立华, 赵智中. 大白菜花茎蜡粉近等基因系转录组分析[J]. 园艺学报, 2022, 49(1): 62-72. |
| [11] | 徐红霞, 周慧芬, 李晓颖, 姜路花, 陈俊伟. 低温胁迫下枇杷不同发育阶段的花果转录组比较分析[J]. 园艺学报, 2021, 48(9): 1680-1694. |
| [12] | 李贵生. 猕猴桃‘金艳'和‘红阳'果实转录组的比较分析[J]. 园艺学报, 2021, 48(6): 1183-1196. |
| [13] | 兰黎明, 罗昌国, 王三红. 基于转录组测序的湖北海棠抗白粉病机制分析[J]. 园艺学报, 2021, 48(5): 860-872. |
| [14] | 鱼尚奇, 张锐, 郭众仲, 宋岩, 付嘉智, 武鹏雨, 马治浩. 核桃内果皮硬化期生长素动态变化及差异表达基因分析[J]. 园艺学报, 2021, 48(3): 487-504. |
| [15] | 庄玥莹, 周利君, 程璧瑄, 于超, 罗乐, 潘会堂, 张启翔. 基于转录组测序的香水月季花香代谢基因研究[J]. 园艺学报, 2021, 48(11): 2262-2274. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司