
园艺学报 ›› 2021, Vol. 48 ›› Issue (4): 719-732.doi: 10.16420/j.issn.0513-353x.2020-0540
收稿日期:2020-12-23
出版日期:2021-04-25
发布日期:2021-04-29
通讯作者:
张俊红
E-mail:zhangjunhng@mail.hzau.edu.cn
基金资助:
LI Guobin, AI Guo, WEI Jing, ZHANG Junhong( )
)
Received:2020-12-23
Online:2021-04-25
Published:2021-04-29
Contact:
ZHANG Junhong
E-mail:zhangjunhng@mail.hzau.edu.cn
摘要:
碱基编辑技术是以CRISPR/Cas系统为基础开发的一种能够对基因组进行定点精准编辑的新技术,包括胞嘧啶碱基编辑系统(cytosine base editor,CBE),腺嘌呤碱基编辑系统(adenine base editor,ABE)以及引导编辑系统(prime editing,PE)。胞嘧啶碱基编辑系统可以将基因组靶位点处的C/G转换为T/A,腺嘌呤碱基编辑系统可以将靶位点处的A/T转变为G/C,而引导编辑系统则可以实现所有12种类型(C-T、G-A、A-G、T-C、C-A、C-G、G-C、G-T、A-C、A-T、T-A、T-G)碱基的任意替换以及碱基的插入和删除。本文中系统介绍了这3种碱基编辑系统的原理、开发过程、各自的优缺点以及在作物遗传改良中的应用和发展,并展望了碱基编辑技术在农作物育种中的应用前景。
中图分类号:
李国斌, 艾国, 韦静, 张俊红. 碱基编辑技术及其在作物遗传改良中的应用综述[J]. 园艺学报, 2021, 48(4): 719-732.
LI Guobin, AI Guo, WEI Jing, ZHANG Junhong. Review of Base Editing Technology and its Application in Crop Genetic Improvement[J]. Acta Horticulturae Sinica, 2021, 48(4): 719-732.
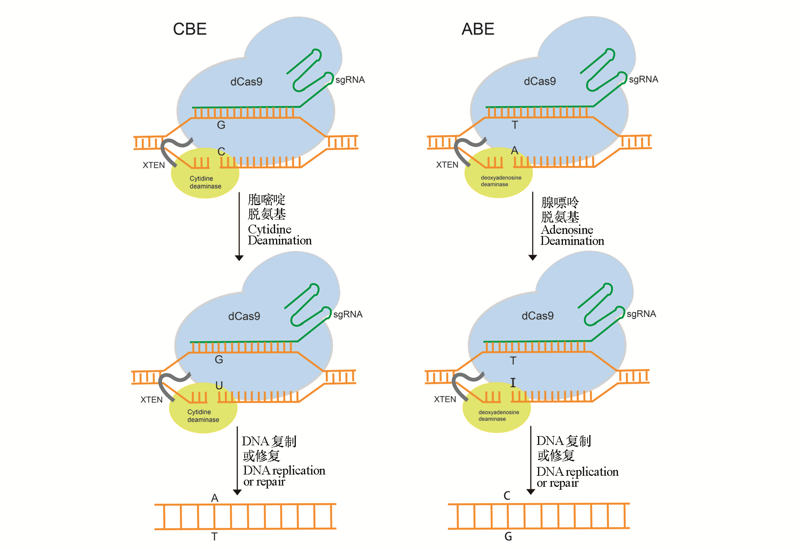
图1 胞嘧啶碱基编辑系统(CBE)和腺嘌呤碱基编辑系统(ABE)示意图
Fig. 1 Schematic diagram of cytosine base editor(CBE)and adenine base editior(ABE) (Komor et al.2016;Gaudelli et al.2017)
| 物种 Crop name | 编辑系统 Base editor | 编辑形式 Editor type | PAM | 靶基因 Targeted gene | 应用 Function | 参考文献 Reference |
|---|---|---|---|---|---|---|
| 水稻Rice | BE3, Anc689BE4max | C-T | NGG,NG | NRT1.1b,SLR1 | 氮利用率、株形 Nitrogen use efficiency,Plant architecture | Lu & Zhu, |
| CBE-P1-P5, ABE-P1-P5 | C-T,A-G | NGG | OsSPL13, OsSPL14, OsSPL16, OsSPL17, OsSPL18, GRF4, OsTOE1, OslDS1, OMTN1, SNB, PMS1, PMS3 | 株形,产量,抗性 Plant architecture,Yield increase,Improve resistance | Hua et al. | |
| ABE-P1S, ABEmax | A-G | NG,NGG | OsSPL14, OsSPL16, OsSERK2, SLR1, Tms9-1, OsNRT1.1B, OsACC1, OsDEP1, OsSPL17, OsSPL18, SLR1, ALS, EPSPS, | 株形,抗除草剂 Plant architecture,Herbicide resistance | Hua et al. | |
| rBE3,rBE4, rBE5,rBE9 | C-T | NGG, NAG, NGA, AGTG, AGCG AGTG, AGCG | OsSERK1, OsSERK2, OsAOS1, OsJAR1, OsJAR2, OsCOL2 | Ren et al. | ||
| rBE14 | A-G | NGG | OsMPK6, OsMPK13, OsSERK2, OsWRKY45, Tms9-1 | 抗病Pathogen resistance | Yan et al. | |
| rBE22, rBE25 | C-T | NG, NAC, NTG, NTT, NCG | OsBZR1 | Ren et al. | ||
| rBE23, rBE26 | A-G | NG, NAC, NTG, NTT, NCG | OsCERK2, OsGS1 | 抗病 Pathogen resistance | Ren et al. | |
| pCXUN-BE3 | C-T | NGG | OsPDS, OsSBEIIb | 品质 Nutritional improvement | Li et al. | |
| A3A-PBE | C-T | NGG, NGA, NGCG, NNGRRT, NNNRRT | OsAAT-T1, OsCDC48, OsDEP1, sNRT1.1B-T1, OsOD, OsEV | Zong et al. | ||
| xCas9-epBE | C-T | GA, NG | OsMPK2, OsMPK5, OsALS, OsNRT1.1B, OsWaxy | 抗病, 抗除草剂 Pathogen resistance, Herbicide resistance | Zhong et al. | |
| 小麦Wheat | A3A-PBE | C-T | NGG, NGA, NGCG, NNGRRT, NNNRRT | TaALS, TaMTL, TaLOX2-T1, TaLOX2-T2 | 抗除草剂 Herbicide resistance | Zong et al. |
| 玉米Maize | PBE(BE3) | C-T | NGG | ZmCENH3 | Zong et al. | |
| 马铃薯 Potato | A3A-PBE | C-T | NGG, NGA, NGCG, NNGRRT, NNNRRT | StGBSS-T6 | Zong et al. | |
| CBE | C-T | NGG | StGBSS1 | 淀粉含量 Starch synthesis | Veillet et al. | |
| 拟南芥 Arabidopsis | BE3 | C-T | NGG | AtALS | 抗除草剂 Herbicide resistance | Chen et al. |
| 番茄Tomato | CBE | C-T | NGG | ALS | 抗除草剂 Herbicide resistance | Veillet et al. |
| Target-AID | C-T | NGG | DELLA, ETR1 | 株型 Plant architecture | Shimatani et al. | |
| 油菜Rape | pcABE7.10 | A-G | NGG | BnALS, BnPDS | 抗除草剂 Herbicide resistance | Kang et al. |
| CBE | C-T | NGG | BnALS1, BnALS3 | 抗除草剂 Herbicide resistance | Wu et al. | |
| 棉花Cotton | GhBE3 | C-T | NGG | GhCAL, GhPEPB | 株型 Plant architecture | Qin et al. |
| 西瓜Watermelon | BE3 | C-T | NGG | ClALS | 抗除草剂 Herbicide resistance | Tian et al. |
表1 单碱基编辑系统在不同作物物中的应用
Table 1 Application of base editors in different crops
| 物种 Crop name | 编辑系统 Base editor | 编辑形式 Editor type | PAM | 靶基因 Targeted gene | 应用 Function | 参考文献 Reference |
|---|---|---|---|---|---|---|
| 水稻Rice | BE3, Anc689BE4max | C-T | NGG,NG | NRT1.1b,SLR1 | 氮利用率、株形 Nitrogen use efficiency,Plant architecture | Lu & Zhu, |
| CBE-P1-P5, ABE-P1-P5 | C-T,A-G | NGG | OsSPL13, OsSPL14, OsSPL16, OsSPL17, OsSPL18, GRF4, OsTOE1, OslDS1, OMTN1, SNB, PMS1, PMS3 | 株形,产量,抗性 Plant architecture,Yield increase,Improve resistance | Hua et al. | |
| ABE-P1S, ABEmax | A-G | NG,NGG | OsSPL14, OsSPL16, OsSERK2, SLR1, Tms9-1, OsNRT1.1B, OsACC1, OsDEP1, OsSPL17, OsSPL18, SLR1, ALS, EPSPS, | 株形,抗除草剂 Plant architecture,Herbicide resistance | Hua et al. | |
| rBE3,rBE4, rBE5,rBE9 | C-T | NGG, NAG, NGA, AGTG, AGCG AGTG, AGCG | OsSERK1, OsSERK2, OsAOS1, OsJAR1, OsJAR2, OsCOL2 | Ren et al. | ||
| rBE14 | A-G | NGG | OsMPK6, OsMPK13, OsSERK2, OsWRKY45, Tms9-1 | 抗病Pathogen resistance | Yan et al. | |
| rBE22, rBE25 | C-T | NG, NAC, NTG, NTT, NCG | OsBZR1 | Ren et al. | ||
| rBE23, rBE26 | A-G | NG, NAC, NTG, NTT, NCG | OsCERK2, OsGS1 | 抗病 Pathogen resistance | Ren et al. | |
| pCXUN-BE3 | C-T | NGG | OsPDS, OsSBEIIb | 品质 Nutritional improvement | Li et al. | |
| A3A-PBE | C-T | NGG, NGA, NGCG, NNGRRT, NNNRRT | OsAAT-T1, OsCDC48, OsDEP1, sNRT1.1B-T1, OsOD, OsEV | Zong et al. | ||
| xCas9-epBE | C-T | GA, NG | OsMPK2, OsMPK5, OsALS, OsNRT1.1B, OsWaxy | 抗病, 抗除草剂 Pathogen resistance, Herbicide resistance | Zhong et al. | |
| 小麦Wheat | A3A-PBE | C-T | NGG, NGA, NGCG, NNGRRT, NNNRRT | TaALS, TaMTL, TaLOX2-T1, TaLOX2-T2 | 抗除草剂 Herbicide resistance | Zong et al. |
| 玉米Maize | PBE(BE3) | C-T | NGG | ZmCENH3 | Zong et al. | |
| 马铃薯 Potato | A3A-PBE | C-T | NGG, NGA, NGCG, NNGRRT, NNNRRT | StGBSS-T6 | Zong et al. | |
| CBE | C-T | NGG | StGBSS1 | 淀粉含量 Starch synthesis | Veillet et al. | |
| 拟南芥 Arabidopsis | BE3 | C-T | NGG | AtALS | 抗除草剂 Herbicide resistance | Chen et al. |
| 番茄Tomato | CBE | C-T | NGG | ALS | 抗除草剂 Herbicide resistance | Veillet et al. |
| Target-AID | C-T | NGG | DELLA, ETR1 | 株型 Plant architecture | Shimatani et al. | |
| 油菜Rape | pcABE7.10 | A-G | NGG | BnALS, BnPDS | 抗除草剂 Herbicide resistance | Kang et al. |
| CBE | C-T | NGG | BnALS1, BnALS3 | 抗除草剂 Herbicide resistance | Wu et al. | |
| 棉花Cotton | GhBE3 | C-T | NGG | GhCAL, GhPEPB | 株型 Plant architecture | Qin et al. |
| 西瓜Watermelon | BE3 | C-T | NGG | ClALS | 抗除草剂 Herbicide resistance | Tian et al. |
| [1] |
Anzalone A V, Randolph P B, Davis J R, Sousa A A, Koblan L W, Levy J M, Chen P J, Wilson C, Newby G A, Raguram A, Liu D R. 2019. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature, 576 (7785):149-157.
doi: 10.1038/s41586-019-1711-4 pmid: 31634902 |
| [2] |
Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero D A, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science, 315 (5819):1709-1712.
doi: 10.1126/science.1138140 URL |
| [3] |
Chatterjee P, Jakimo N, Jacobson J M. 2018. Minimal PAM specificity of a highly similar SpCas9 ortholog. Science Advances, 4 (10):eaau0766.
doi: 10.1126/sciadv.aau0766 URL |
| [4] |
Chen Y, Wang Z, Ni H, Xu Y, Chen Q, Jiang L. 2017. CRISPR/Cas9-mediated base-editing system efficiently generates gain-of-function mutations in Arabidopsis. Science China Life Sciences, 60 (5):520-523.
doi: 10.1007/s11427-017-9021-5 URL |
| [5] |
Chen Y T, Mao W W, Liu T, Feng Q Q, Li L, Li B B. 2020. Genome editing as a versatile tool to improve horticultural crop qualities. Horticultural Plant Journal, 6 (6):372-384.
doi: 10.1016/j.hpj.2020.11.004 URL |
| [6] | Cui Xia,Zhang Shuaibin. 2017. The utilization and prospect of genome editing in horticultural crops. Acta Horticulturae Sinica, 44 (9):1787-1795. (in Chinese) |
| 崔霞, 张率斌. 2017. 基因编辑技术及其在园艺作物中的应用和展望. 园艺学报, 44 (9):1787-1795. | |
| [7] |
d'Adda di Fagagna F, Weller G R, Doherty A J, Jackson S P. 2003. The Gam protein of bacteriophage Mu is an orthologue of eukaryotic Ku. EMBO Reports, 4 (1):47-52.
doi: 10.1038/sj.embor.embor709 URL |
| [8] |
Gaudelli N M, Komor A C, Rees H A, Packer M S, Badran A H, Bryson D I, Liu D R. 2017. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature, 551 (7681):464-471.
doi: 10.1038/nature24644 pmid: 29160308 |
| [9] |
Horvath P, Barrangou R. 2010. CRISPR/Cas,the immune system of bacteria and archaea. Science, 327 (5962):167-170.
doi: 10.1126/science.1179555 URL |
| [10] |
Hua K, Jiang Y, Tao X, Zhu J K. 2020a. Precision genome engineering in rice using prime editing system. Plant Biotechnology Journal, 18 (11):2167-2169.
doi: 10.1111/pbi.v18.11 URL |
| [11] |
Hua K, Tao X, Liang W, Zhang Z, Gou R, Zhu J K. 2020b. Simplified adenine base editors improve adenine base editing efficiency in rice. Plant Biotechnology Journal, 18 (3):770-778.
doi: 10.1111/pbi.v18.3 URL |
| [12] |
Hua K, Tao X, Yuan F, Wang D, Zhu J K. 2018. Precise A.T to G.C base editing in the rice genome. Molecular Plant, 11 (4):627-630.
doi: 10.1016/j.molp.2018.02.007 URL |
| [13] |
Hua K, Tao X, Zhu J K. 2019. Expanding the base editing scope in rice by using Cas9 variants. Plant Biotechnology Journal, 17 (2):499-504.
doi: 10.1111/pbi.2019.17.issue-2 URL |
| [14] | Jin S, Zong Y, Wang D, Gao Q, Qiu J, Zhu Z, Zhang F, Gao C. 2019. Cytosine,but not adenine,base editors induce genome-wide off-target mutations in rice. Science, 364 (6347):292-295. |
| [15] |
Kang B C, Yun J Y, Kim S T, Shin Y, Ryu J, Choi M, Woo J W, Kim J S. 2018. Precision genome engineering through adenine base editing in plants. Nature Plants, 4 (7):427-431.
doi: 10.1038/s41477-018-0178-x URL |
| [16] |
Keiji Nishida, Masao Mochizuki, Takayuki Arazoe, Aya Miyabe, Nozomu Yachie, Michihiro Araki, Satomi Banno, Mika Kakimoto, Kiyotaka Y. Hara, Mayura Tabata, Zenpei Shimatani, Kondo A. . 2016. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science, 353 (6305):aaf8729.
doi: 10.1126/science.aaf8729 URL |
| [17] |
Kim Y B, Komor A C, Levy J M, Packer M S, Zhao K T, Liu D R. 2017. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nature Biotechnology, 35 (4):371-376.
doi: 10.1038/nbt.3803 URL |
| [18] |
Komor A C, Kevin T Zhao, Michael S Packer, Nicole M Gaudelli, Amanda L Waterbury, Luke W Koblan, Y. Bill Kim, Ahmed H Badran, Liu D R. 2017. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C_G-to-T_A base editors with higher efficiency and product purity. Science Advances, 3 (8):eaao4774.
doi: 10.1126/sciadv.aao4774 URL |
| [19] |
Komor A C, Kim Y B, Packer M S, Zuris J A, Liu D R. 2016. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature, 533 (7603):420-424.
doi: 10.1038/nature17946 URL |
| [20] |
Kuang Y, Li S, Ren B, Yan F, Spetz C, Li X, Zhou X, Zhou H. 2020. Base-editing-mediated artificial evolution of OsALS 1 in planta to develop novel herbicide-tolerant rice germplasms. Molecular Plant, 13 (4):565-572.
doi: 10.1016/j.molp.2020.01.010 URL |
| [21] |
Landrum M J, Lee J M, Benson M, Brown G, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Hoover J, Jang W, Katz K, Ovetsky M, Riley G, Sethi A, Tully R, Villamarin-Salomon R, Rubinstein W, Maglott D R. 2016. ClinVar:public archive of interpretations of clinically relevant variants. Nucleic Acids Research, 44 (D1):D862-868.
doi: 10.1093/nar/gkv1222 URL |
| [22] | Li C, Zhang R, Meng X, Chen S, Zong Y, Lu C, Qiu J L, Chen Y H, Li J, Gao C. 2020a. Targeted,random mutagenesis of plant genes with dual cytosine and adenine base editors. Nature biotechnology, 38 (7):U875-U866. |
| [23] |
Li C, Zong Y, Wang Y, Jin S, Zhang D, Song Q, Zhang R, Gao C. 2018a. Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biology, 19 (1):19-59.
doi: 10.1186/s13059-018-1398-0 URL |
| [24] |
Li H, Li J, Chen J, Yan L, Xia L. 2020b. Precise modifications of both exogenous and endogenous genes in rice by prime editing. Molecular Plant, 13 (5):671-674.
doi: 10.1016/j.molp.2020.03.011 URL |
| [25] |
Li J, Sun Y, Du J, Zhao Y, Xia L. 2017. Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system. Molecular Plant, 10 (3):526-529.
doi: 10.1016/j.molp.2016.12.001 URL |
| [26] | Li J, Zhang Y, Chen K L, Shan Q W, Wang Y P, Liang Z, Gao C X. 2013. CRISPR/Cas:a novel way of RNA-guided genome editing. Hereditas, 35 (11):1265-1273. |
| [27] |
Li X, Wang Y, Liu Y, Yang B, Wang X, Wei J, Lu Z, Zhang Y, Wu J, Huang X, Yang L, Chen J. 2018b. Base editing with a Cpf1-cytidine deaminase fusion. Nature Biotechnology, 36 (4):324-327.
doi: 10.1038/nbt.4102 URL |
| [28] |
Li Z, Xiong X, Wang F, Liang J, Li J F. 2019. Gene disruption through base editing-induced messenger RNA missplicing in plants. The New Phytologist, 222 (2):1139-1148.
doi: 10.1111/nph.2019.222.issue-2 URL |
| [29] |
Lin Q, Zong Y, Xue C, Wang S, Jin S, Zhu Z, Wang Y, Anzalone A V, Raguram A, Doman J L, Liu D R, Gao C. 2020. Prime genome editing in rice and wheat. Nature Biotechnology, 38 (5):582-585.
doi: 10.1038/s41587-020-0455-x URL |
| [30] | Liu Jiahui, Liang Puping, Shi Guang, Huang Junjiu. 2017. Research progress of base editing system. World SCI-TECH R & D, 39 (6):457-462. (in Chinese) |
| 刘佳慧, 梁普平, 时光, 黄军就. 2017. 单碱基基因编辑系统的研究进展. 世界科技研究与发展, 39 (6):457-462. | |
| [31] |
Lu Y, Zhu J K. 2017. Precise editing of a target base in the rice genome using a modified CRISPR/Cas9 system. Molecular Plant, 10 (3):523-525.
doi: 10.1016/j.molp.2016.11.013 URL |
| [32] |
Ma C F, Liu M C, Li Q F, Si J, Ren X S, Song H Y. 2019. Efficient BoPDS gene editing in cabbage by the CRISPR/Cas9 system. Horticultural Plant Journal, 5 (4):164-169.
doi: 10.1016/j.hpj.2019.04.001 URL |
| [33] |
Ma Y, Zhang J, Yin W, Zhang Z, Song Y, Chang X. 2016. Targeted AID-mediated mutagenesis(TAM)enables efficient genomic diversification in mammalian cells. Nature Methods, 13 (12):1029-1035.
doi: 10.1038/nmeth.4027 URL |
| [34] |
Mishra R, Joshi R K, Zhao K. 2020. Base editing in crops:current advances,limitations and future implications. Plant Biotechnology Journal, 18 (1):20-31.
doi: 10.1111/pbi.v18.1 URL |
| [35] |
Nishimasu H, Shi X, Ishiguro S, Gao L, Hirano S, Okazaki S, Noda T, Abudayyeh O O, Gootenberg J S, Mori H, Oura S, Holmes B, Tanaka M, Seki M, Hirano H, Aburatani H, Ishitani R, Ikawa M, Yachie N, Zhang F, Nureki O. 2018. Engineered CRISPR-Cas 9 nuclease with expanded targeting space. Science, 361 (6408):1259-1262.
doi: 10.1126/science.aas9129 URL |
| [36] |
Qin L, Li J, Wang Q, Xu Z, Sun L, Alariqi M, Manghwar H, Wang G, Li B, Ding X, Rui H, Huang H, Lu T, Lindsey K, Daniell H, Zhang X, Jin S. 2020. High-efficient and precise base editing of C*G to T*A in the allotetraploid cotton(Gossypium hirsutum)genome using a modified CRISPR/Cas9 system. Plant Biotechnology Journal, 18 (1):45-56.
doi: 10.1111/pbi.v18.1 URL |
| [37] |
Rees H A, Komor A C, Yeh W H, Caetano-Lopes J, Warman M, Edge A S B, Liu D R. 2017. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nature Communications, 8:15790.
doi: 10.1038/ncomms15790 URL |
| [38] |
Ren B, Liu L, Li S, Kuang Y, Wang J, Zhang D, Zhou X, Lin H, Zhou H. 2019. Cas9-NG greatly expands the targeting scope of the genome-editing toolkit by recognizing NG and other atypical PAMs in rice. Molecular Plant, 12 (7):1015-1026.
doi: 10.1016/j.molp.2019.03.010 URL |
| [39] |
Ren B, Yan F, Kuang Y, Li N, Zhang D, Lin H, Zhou H. 2017. A CRISPR/Cas 9 toolkit for efficient targeted base editing to induce genetic variations in rice. Science China Life Sciences, 60 (5):516-519.
doi: 10.1007/s11427-016-0406-x URL |
| [40] |
Ren B, Yan F, Kuang Y, Li N, Zhang D, Zhou X, Lin H, Zhou H. 2018. Improved base editor for efficiently inducing genetic variations in rice with CRISPR/Cas9-guided hyperactive hAID mutant. Molecular Plant, 11 (4):623-626.
doi: 10.1016/j.molp.2018.01.005 URL |
| [41] |
Satomura A, Nishioka R, Mori H, Sato K, Kuroda K, Ueda M. 2017. Precise genome-wide base editing by the CRISPR Nickase system in yeast. Scientific Reports, 7 (1):2095.
doi: 10.1038/s41598-017-02013-7 pmid: 28522803 |
| [42] |
Schellenberger V, Wang C W, Geething N C, Spink B J, Campbell A, To W, Scholle M D, Yin Y, Yao Y, Bogin O, Cleland J L, Silverman J, Stemmer W P. 2009. A recombinant polypeptide extends the in vivo half-life of peptides and proteins in a tunable manner. Nature Biotechnology, 27 (12):1186-1190.
doi: 10.1038/nbt.1588 pmid: 19915550 |
| [43] |
Shimatani Z, Kashojiya S, Takayama M, Terada R, Arazoe T, Ishii H, Teramura H, Yamamoto T, Komatsu H, Miura K, Ezura H, Nishida K, Ariizumi T, Kondo A. 2017. Targeted base editing in rice and tomato using a CRISPR-Cas 9 cytidine deaminase fusion. Nature Biotechnology, 35 (5):441-443.
doi: 10.1038/nbt.3833 pmid: 28346401 |
| [44] |
Swanton C, McGranahan N, Starrett G J, Harris R S. 2015. APOBEC Enzymes:mutagenic fuel for cancer evolution and heterogeneity. Cancer Discovery, 5 (7):704-712.
doi: 10.1158/2159-8290.CD-15-0344 URL |
| [45] |
Tang X, Sretenovic S, Ren Q, Jia X, Li M, Fan T, Yin D, Xiang S, Guo Y, Liu L, Zheng X, Qi Y, Zhang Y. 2020. Plant prime editors enable precise gene editing in rice cells. Molecular Plant, 13 (5):667-670.
doi: S1674-2052(20)30072-1 pmid: 32222487 |
| [46] |
Tian S, Jiang L, Cui X, Zhang J, Guo S, Li M, Zhang H, Ren Y, Gong G, Zong M, Liu F, Chen Q, Xu Y. 2018. Engineering herbicide-resistant watermelon variety through CRISPR/Cas9-mediated base-editing. Plant Cell Reports, 37 (9):1353-1356.
doi: 10.1007/s00299-018-2299-0 URL |
| [47] |
Veillet F, Chauvin L, Kermarrec M P, Sevestre F, Merrer M, Terret Z, Szydlowski N, Devaux P, Gallois J L, Chauvin J E. 2019a. The Solanum tuberosum GBSSI gene:a target for assessing gene and base editing in tetraploid potato. Plant Cell Reports, 38 (9):1065-1080.
doi: 10.1007/s00299-019-02426-w URL |
| [48] |
Veillet F, Perrot L, Chauvin L, Kermarrec M P, Guyon-Debast A, Chauvin J E, Nogue F, Mazier M. 2019b. Transgene-free genome editing in tomato and potato plants using Agrobacterium-mediated delivery of a CRISPR/Cas 9 cytidine base editor. International Journal of Molecular Sciences, 20 (2):402.
doi: 10.3390/ijms20020402 URL |
| [49] |
Wang M, Wang Z, Mao Y, Lu Y, Yang R, Tao X, Zhu J K. 2019. Optimizing base editors for improved efficiency and expanded editing scope in rice. Plant Biotechnology Journal, 17 (9):1697-1699.
doi: 10.1111/pbi.v17.9 URL |
| [50] |
Wang M, Xu Z, Gosavi G, Ren B, Cao Y, Kuang Y, Zhou C, Spetz C, Yan F, Zhou X, Zhou H. 2020. Targeted base editing in rice with CRISPR/ScCas9 system. Plant Biotechnology Journal, 18 (8):1645-1647.
doi: 10.1111/pbi.v18.8 URL |
| [51] |
Wu J, Chen C, Xian G, Liu D, Lin L, Yin S, Sun Q, Fang Y, Zhang H, Wang Y. 2020. Engineering herbicide-resistant oilseed rape by CRISPR/Cas9-mediated cytosine base-editing. Plant Biotechnology Journal, 18 (9):1857-1859.
doi: 10.1111/pbi.v18.9 URL |
| [52] |
Xu R, Li J, Liu X, Shan T, Qin R, Wei P. 2020a. Development of plant prime-editing systems for precise genome editing. Plant Communications, 1 (3):100043.
doi: 10.1016/j.xplc.2020.100043 URL |
| [53] |
Xu W, Zhang C, Yang Y, Zhao S, Kang G, He X, Song J, Yang J. 2020b. Versatile nucleotides substitution in plant using an improved prime editing system. Molecular Plant, 13 (5):675-678.
doi: 10.1016/j.molp.2020.03.012 URL |
| [54] |
Xue C, Zhang H, Lin Q, Fan R, Gao C. 2018. Manipulating mRNA splicing by base editing in plants. Science China Life Sciences, 61 (11):1293-1300.
doi: 10.1007/s11427-018-9392-7 URL |
| [55] |
Yan F, Kuang Y, Ren B, Wang J, Zhang D, Lin H, Yang B, Zhou X, Zhou H. 2018. Highly efficient A.T to G.C base editing by Cas9n-Guided tRNA adenosine deaminase in rice. Molecular Plant, 11 (4):631-634.
doi: 10.1016/j.molp.2018.02.008 URL |
| [56] |
Yang B, Yang L, Chen J. 2019. Development and application of base editors. The CRISPR Journal, 2 (2):91-104.
doi: 10.1089/crispr.2019.0001 pmid: 30998092 |
| [57] |
Yasui M, Suenaga E, Koyama N, Masutani C, Hanaoka F, Gruz P, Shibutani S, Nohmi T, Hayashi M, Honma M. 2008. Miscoding properties of 2'-deoxyinosine,a nitric oxide-derived DNA Adduct,during translesion synthesis catalyzed by human DNA polymerases. Journal of Molecular Biology, 377 (4):1015-1023.
doi: 10.1016/j.jmb.2008.01.033 URL |
| [58] |
Zhang C, Xu W, Wang F, Kang G, Yuan S, Lv X, Li L, Liu Y, Yang J. 2020. Expanding the base editing scope to GA and relaxed NG PAM sites by improved xCas9 system. Plant Biotechnology Journal, 18 (4):884-886.
doi: 10.1111/pbi.v18.4 URL |
| [59] | Zhao K, Tung C W, Eizenga G C, Wright M H, Ali M L, Price A H, Norton G J, Islam M R, Reynolds A, Mezey J, McClung A M, Bustamante C D, McCouch S R. 2011. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nature Communications, 2 (167):1-10. |
| [60] |
Zhong Z, Sretenovic S, Ren Q, Yang L, Bao Y, Qi C, Yuan M, He Y, Liu S, Liu X, Wang J, Huang L, Wang Y, Baby D, Wang D, Zhang T, Qi Y, Zhang Y. 2019. Improving plant genome editing with high-fidelity xCas9 and non-canonical PAM-targeting Cas9-NG. Molecular Plant, 12 (7):1027-1036.
doi: 10.1016/j.molp.2019.03.011 URL |
| [61] |
Zong Y, Song Q, Li C, Jin S, Zhang D, Wang Y, Qiu J L, Gao C. 2018. Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nature Biotechnology, 36 (10)
doi: 10.1038/nbt0118-10 URL |
| [62] |
Zong Y, Wang Y, Li C, Zhang R, Chen K, Ran Y, Qiu J L, Wang D, Gao C. 2017. Precise base editing in rice,wheat and maize with a Cas9-cytidine deaminase fusion. Nature Biotechnology, 35 (5):438-440.
doi: 10.1038/nbt.3811 URL |
| [63] | Zong Yuan, Gao Caixia. 2019. Progress on base editing systems. Hereditas, 41 (9):777-800. (in Chinese) |
| 宗媛, 高彩霞. 2019. 碱基编辑系统研究进展. 遗传, 41 (9):777-800. |
| [1] | 左鑫, 李铭铭, 李欣容, 苗春妍, 李炎枋, 杨旭, 张重义, 王丰青. CRISPR/Cas9技术在天目地黄RcPDS1基因编辑中的应用[J]. 园艺学报, 2022, 49(7): 1532-1544. |
| [2] | 贾芝琪;崔艳红;李颖;杨宇红;黄三文;杜永臣. 马铃薯抗晚疫病基因R3a、R1和RB在番茄中的表达[J]. 园艺学报, 2009, 36(8): 1153-1160. |
| [3] | 叶庆亮;江 东;彭爱红. ‘岩溪晚芦’椪柑果皮与果肉差减cDNA文库的构建及初步分析[J]. 园艺学报, 2009, 36(7): 967-974. |
| [4] | 肖 罗;陈国华;戴良英;吕春花;杨宇红;谢丙炎. 香蕉穿孔线虫β-1,4-内切葡聚糖酶基因的克隆与特征分析[J]. 园艺学报, 2008, 35(10): 1431-1440. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司