
园艺学报 ›› 2021, Vol. 48 ›› Issue (4): 632-646.doi: 10.16420/j.issn.0513-353x.2020-0506
收稿日期:2020-12-10
出版日期:2021-04-25
发布日期:2021-04-29
通讯作者:
齐红岩
E-mail:qihongyan@syau.edu.cn
基金资助:
YUE Lingqi, XING Qiaojuan, ZHANG Xiaolan, LIANG Xue, WANG Qian, QI Hongyan( )
)
Received:2020-12-10
Online:2021-04-25
Published:2021-04-29
Contact:
QI Hongyan
E-mail:qihongyan@syau.edu.cn
摘要:
光敏色素互作因子(Phytochrome-interacting factor,PIF)属于碱性螺旋—环—螺旋(Basic Helix-Loop-Helix,bHLH)类转录因子,参与植物多个生物进程,作为胞内信号调节“枢纽”,不但参与调控植物的生长发育,且在植物抵御逆境胁迫过程中发挥重要作用。本文中主要阐述PIF集成复杂的网络调控植物对低温、高温、荫蔽、干旱等非生物胁迫的应答反应,以及PIF通过植物激素信号途径介导植物对病虫害等生物胁迫的防御机制,为研究PIF在逆境胁迫中的作用,提高作物抗性和品质等提供参考。
中图分类号:
岳玲琦, 邢巧娟, 张晓兰, 梁雪, 王乾, 齐红岩. 光敏色素互作因子在植物抵御逆境胁迫中的作用研究进展[J]. 园艺学报, 2021, 48(4): 632-646.
YUE Lingqi, XING Qiaojuan, ZHANG Xiaolan, LIANG Xue, WANG Qian, QI Hongyan. Research Progress on the Effect of Phytochrome-interacting Factors in Plant Resistance to Abiotic Stress[J]. Acta Horticulturae Sinica, 2021, 48(4): 632-646.
| 物种 Species | PIF数量 PIF quantity | 含APB结构域成员 Members that contains the APB domain | 含APA结构域成员 Members that contains the APA domain | 参考文献 Reference |
|---|---|---|---|---|
| 拟南芥Arabidopsis thaliana | 8 | AtPIF1 ~ AtPIF8 | AtPIF1,AtPIF3 | Leivar & Quail, Lee & Choi, |
| 水稻Oryza sativa | 6 | OsPIL11 ~ OsPIL16 | OsPIL15 | Cordeiro et al. |
| 玉米Zea mays | 7 | ZmbHLH002 ~ ZmbHLH008 | ZmbHLH005(ZmPIF3.1), ZmbHLH004(ZmPIF3.2) | Kumar et al. |
| 小麦Triticum aestivum | 1 | — | — | 李璐 等, |
| 番茄Lycopersicon esculentum | 8 | SlPIF1a,SlPIF1b,SlPIF3,SlPIF4,SlPIF7a,SlPIF7b,SlPIF8a,SlPIF8b | SlPIF1a,SlPIF1b,SlPIF3 | Rosado et al. |
| 苹果Malus×domestica | 4 | MdPIF1,MdPIF3 MdPIF5,MdPIF7 | — | 宋杨 等, 谯愚, |
| 草莓Fragaria × ananassa | 1 | — | — | 李瑞玲 等, |
| 葡萄Vitis vinifera | 4 | VvPIF3,VvPIF4,VvPIF7 | VvPIF1,VvPIF3 | Zhang et al. |
| 甘蓝型油菜Brassica napus | 1 | — | — | 冯韬和官春云, |
| 毛果杨Populus trichocarpa | 10 | — | — | 徐向东 等, |
| 小立碗藓Physcomitrella patens | 4 | PpPIF1 ~ PpPIF4(含APB-like) | PpPIF1-PpPIF4 | Possart et al. |
| 地钱Marchantia polymorpha | 1 | — | MpPIF(含推测的APA) | Inoue et al. |
表1 植物中PIF家族成员数量及其氨基酸序列含有的特殊结构域
Table 1 Number of members of the PIF genes family in plants and the specific structural domains contained in their protein sequences
| 物种 Species | PIF数量 PIF quantity | 含APB结构域成员 Members that contains the APB domain | 含APA结构域成员 Members that contains the APA domain | 参考文献 Reference |
|---|---|---|---|---|
| 拟南芥Arabidopsis thaliana | 8 | AtPIF1 ~ AtPIF8 | AtPIF1,AtPIF3 | Leivar & Quail, Lee & Choi, |
| 水稻Oryza sativa | 6 | OsPIL11 ~ OsPIL16 | OsPIL15 | Cordeiro et al. |
| 玉米Zea mays | 7 | ZmbHLH002 ~ ZmbHLH008 | ZmbHLH005(ZmPIF3.1), ZmbHLH004(ZmPIF3.2) | Kumar et al. |
| 小麦Triticum aestivum | 1 | — | — | 李璐 等, |
| 番茄Lycopersicon esculentum | 8 | SlPIF1a,SlPIF1b,SlPIF3,SlPIF4,SlPIF7a,SlPIF7b,SlPIF8a,SlPIF8b | SlPIF1a,SlPIF1b,SlPIF3 | Rosado et al. |
| 苹果Malus×domestica | 4 | MdPIF1,MdPIF3 MdPIF5,MdPIF7 | — | 宋杨 等, 谯愚, |
| 草莓Fragaria × ananassa | 1 | — | — | 李瑞玲 等, |
| 葡萄Vitis vinifera | 4 | VvPIF3,VvPIF4,VvPIF7 | VvPIF1,VvPIF3 | Zhang et al. |
| 甘蓝型油菜Brassica napus | 1 | — | — | 冯韬和官春云, |
| 毛果杨Populus trichocarpa | 10 | — | — | 徐向东 等, |
| 小立碗藓Physcomitrella patens | 4 | PpPIF1 ~ PpPIF4(含APB-like) | PpPIF1-PpPIF4 | Possart et al. |
| 地钱Marchantia polymorpha | 1 | — | MpPIF(含推测的APA) | Inoue et al. |
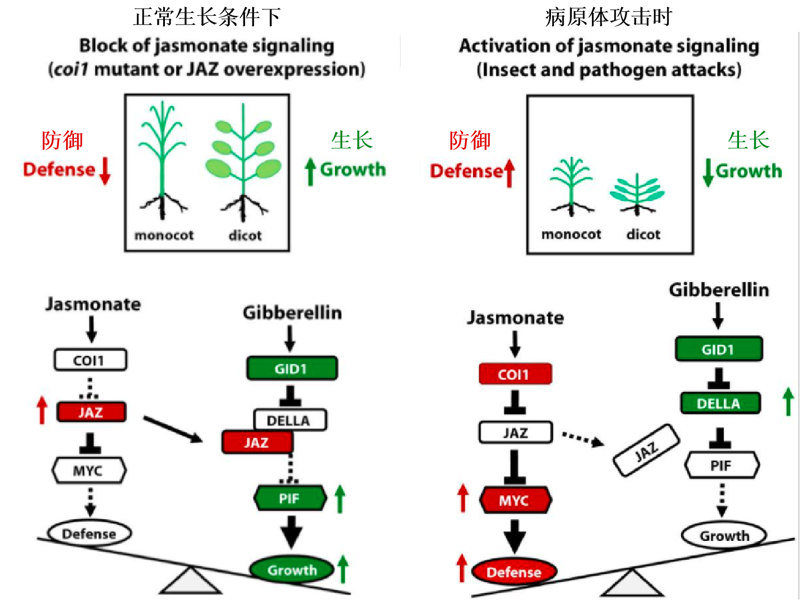
图2 COI1-JAZ-DELLA-PIF平衡植物生长与防御反应的模式图 引自Yang等(2012),略作修改。
Fig. 2 Pattern of COI1-JAZ-DELLA-PIF balancing plant growth and defense response Quoted by Yang et al.2012),with minor modifications.
| [1] |
Achard P, Gusti A, Cheminant S, Alioua M, Dhondt S, Coppens F, Beemster G T S, Genschik P. 2009. Gibberellin signaling controls cell proliferation rate in Arabidopsis. Current Biology, 19 (14):1188-1193.
doi: 10.1016/j.cub.2009.05.059 URL |
| [2] |
Al-Sady B, Ni W, Kircher S, Schafer E, Quail P H. 2006. Photoactivated phytochrome induces rapid PIF 3 phosphorylation prior to proteasome-mediated degradation. Molecular Cell, 23 (3):439-446.
doi: 10.1016/j.molcel.2006.06.011 URL |
| [3] |
Balcerowicz M. 2020. PHYTOCHROME-INTERACTING FACTORS at the interface of light and temperature signalling. Physiologia Plantarum, 169 (3):347-356.
doi: 10.1111/ppl.v169.3 URL |
| [4] | Ballare C L, Pierik R. 2017. The shade-avoidance syndrome:multiple signals and ecological consequences. Plant Cell & Environment, 40 (11):2530-2543. |
| [5] |
Bernardo-Garcia S, de Lucas M, Martinez C, Espinosa-Ruiz A, Daviere J M, Prat S. 2014. BR-dependent phosphorylation modulates PIF 4 transcriptional activity and shapes diurnal hypocotyl growth. Genes & Development, 28 (15):1681-1694.
doi: 10.1101/gad.243675.114 URL |
| [6] |
Bu Q, Zhu L, Dennis M D, Yu L, Lu S X, Person M D, Tobin E M, Browning K S, Huq E. 2011. Phosphorylation by CK2 enhances the rapid light-induced degradation of Phytochrome Interacting Factor 1 in Arabidopsis. Journal of Biological Chemistry, 286 (14):12066-12074.
doi: 10.1074/jbc.M110.186882 URL |
| [7] |
Campos M L, Yoshida Y, Major I T, Ferreira D D O, Weraduwage S M, Froehlich J E, Johnson B F, Kramer D M, Jander G, Sharkey T D, Howe G A. 2016. Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nature Communications, 7:12570.
doi: 10.1038/ncomms12570 URL |
| [8] | Casal J J. 2012. Shade avoidance. The Arabidopsis book. American Society of Plant Biologists, 10:e0157. |
| [9] | Casson S A, Franklin K A, Gray J E, Grierson C S, Whitelam G C, Hetherington A M. 2009. Phytochrome B and PIF 4 regulate stomatal development in response to light quantity. Comparative biochemistry and physiology part A:Molecular & Integrative Physiology, 19 (3):229. |
| [10] | Chang Bo-wen. 2018. Cloning and analysis of genes related to potato shade-resistance response[M. D. Dissertation]. Hefei:Anhui Agricultural University. (in Chinese) |
| 常博雯. 2018. 马铃薯避荫反应相关基因的克隆与分析[硕士论文]. 合肥:安徽农业大学. | |
| [11] | Chen Xiao-xiao. 2019. The mechanisms of PIF 4 mediated light quality-regulated cold tolerance in tomato[M. D. Dissertation]. Hangzhou:Zhejiang University. (in Chinese) |
| 陈笑笑. 2019. PIF4介导光质调控番茄低温抗性的作用机制[硕士论文]. 杭州:浙江大学. | |
| [12] | Cordeiro A M, Figueiredo D D, Tepperman J, Borba A R, Lourenco T, Abreu I A, Ouwerkerk P B F, Quail P H, Oliveira M M, Saibo N J M. 2016. Rice phytochrome-interacting factor protein OsPIF14 represses OsDREB1B gene expression through an extended N-box and interacts preferentially with the active form of phytochrome B. Biochimica et Biophysica Acta(BBA)-Gene Regulatory Mechanisms, 1859:393-404. |
| [13] |
de Lucas M, Daviere J M, Rodriguez-Falcon M, Pontin M, Iglesias-Pedraz J M, Lorrain S, Fankhauser C, Blazquez M A, Titarenko E, Prat S. 2008. A molecular framework for light and gibberellin control of cell elongation. Nature, 451 (7177):480-484.
doi: 10.1038/nature06520 URL |
| [14] |
de Lucas M, Prat S. 2014. PIFs get BRright:PHYTOCHROME INTERACTING FACTORs as integrators of light and hormonal signals. New Phytologist, 202 (4):1126-1141.
doi: 10.1111/nph.2014.202.issue-4 URL |
| [15] |
de Wit M, Keuskamp D H, Bongers F J, Hornitschek P, Gommers C M M, Reinen E, Martinez-Ceron C, Fankhauser C, Pierik R. 2016. Integration of phytochrome and cryptochrome signals determines plant growth during competition for light. Current Biology, 26 (24):3320-3326.
doi: 10.1016/j.cub.2016.10.031 URL |
| [16] |
Dong J, Ni W, Yu R, Deng X W, Chen H, Wei N. 2017. Light-dependent degradation of PIF3 by SCFEBF1/2 promotes a photomorphogenic response in Arabidopsis. Current Biology, 27:2420-2430.
doi: 10.1016/j.cub.2017.06.062 URL |
| [17] |
Dong J, Tang D, Gao Z, Yu R, Li K, He H, Terzaghi W, Deng X W, Chen H. 2014. Arabidopsis DE-ETIOLATED 1 represses photomorphogenesis by positively regulating phytochrome-interacting factors in the dark. Plant Cell, 26 (9):3630-3645.
doi: 10.1105/tpc.114.130666 URL |
| [18] |
Eriksson S, Bohlenius H, Moritz T, Nilsson O. 2006. GA 4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell, 18 (9):2172-2181.
pmid: 16920780 |
| [19] |
Feng Tao, Guan Chun-yun. 2019. Cloning and characterization of phytochrome interacting factor 4 (BnaPIF4) gene from Brassica napus L. Acta Agronomica Sinica, 45 (2):204-213. (in Chinese)
doi: 10.3724/SP.J.1006.2019.84085 URL |
| 冯韬, 官春云. 2019. 甘蓝型油菜光敏色素互作因子4(BnaPIF4)基因克隆和功能分析. 作物学报, 45 (2):204-213. | |
| [20] |
Fernandez V, Takahashi Y, Le Gourrierec J, Coupland G. 2016. Photoperiodic and thermosensory pathways interact through CONSTANS to promote flowering at high temperature under short days. Plant Journal, 86 (5):426-440.
doi: 10.1111/tpj.2016.86.issue-5 URL |
| [21] |
Fiorucci A S, Galvao V C, Ince Y C, Boccaccini A, Goyal A, Allenbach Petrolati L, Trevisan M, Fankhauser C. 2020. PHYTOCHROME INTERACTING FACTOR 7 is important for early responses to elevated temperature in Arabidopsis seedlings. New Phytologist, 226 (1):50-58.
doi: 10.1111/nph.v226.1 URL |
| [22] |
Gangappa S N, Berriri S, Kumar S V. 2017. PIF 4 Coordinates thermosensory growth and immunity in Arabidopsis. Current Biology, 27 (2):243-249.
doi: 10.1016/j.cub.2016.11.012 URL |
| [23] |
Gangappa S N, Kumar S V. 2018. DET1 and COP 1 modulate the coordination of growth and immunity in response to key seasonal signals in Arabidopsis. Cell Reports, 25 (1):29-37.
doi: 10.1016/j.celrep.2018.08.096 URL |
| [24] | Gao Y, Jiang W, Dai Y, Xiao N, Zhang C, Li H, Lu Y, Wu M, Tao X, Deng D, Chen J. 2015. A maize phytochrome-interacting factor 3 improves drought and salt stress tolerance in rice. Plant Molecular Biology, 87 (4-5):413-428. |
| [25] | Gao Yong, Ren Xiao-yun, Li Qian, Qian Jing-jie, Dong Ding, Chen Jian-min. 2019. The drought-resistant function of the transcription factor PIF 3 in Arabidopsis. Journal of Yangzhou University(Agricultural and Life Science Edition), 40 (1):8-13. (in Chinese) |
| 高勇, 任小芸, 李倩, 钱静洁, 董锭, 陈建民. 2019. 拟南芥光敏色素作用因子PIF3节水抗旱功能分析. 扬州大学学报(农业与生命科学版), 40 (1):8-13. | |
| [26] |
Hayes S, Pantazopoulou C K, van Gelderen K, Reinen E, Tween A L, Sharma A, de Vries M, Prat S, Schuurink R C, Testerink C, Pierik R. 2019. Soil salinity limits plant shade avoidance. Current Biology, 29 (10):1669-1676.
doi: 10.1016/j.cub.2019.03.042 URL |
| [27] |
Hoecker U. 2017. The activities of the E3 ubiquitin ligase COP1/SPA,a key repressor in light signaling. Current Opinion in Plant Biology, 37:63-69.
doi: S1369-5266(16)30196-0 pmid: 28433946 |
| [28] |
Horvath D P, Hansen S A, Moriles-Miller J P, Pierik R, Yan C, Clay D E, Scheffler B, Clay S A. 2015. RNAseq reveals weed-induced PIF3-like as a candidate target to manipulate weed stress response in soybean. New Phytologist, 207 (1):196-210.
doi: 10.1111/nph.2015.207.issue-1 URL |
| [29] |
Huai J, Zhang X, Li J, Ma T, Zha P, Jing Y, Lin R. 2018. SEUSS and PIF4 coordinately regulate light and temperature signaling pathways to control plant growth. Molecular Plant, 11:928-942.
doi: 10.1016/j.molp.2018.04.005 URL |
| [30] |
Huang W F, Hu B, Liu J L, Zhou Y, Liu S Q. 2020. Identification and characterization of tonoplast sugar transporter(TST)gene family in cucumber. Horticultural Plant Journal, 6 (3):145-157.
doi: 10.1016/j.hpj.2020.03.005 URL |
| [31] |
Huot B, Yao J, Montgomery B L, He S Y. 2014. Growth-defense tradeoffs in plants:a balancing act to optimize fitness. Molecular Plant, 7 (8):1267-1287.
doi: 10.1093/mp/ssu049 URL |
| [32] | Huq E, Al-Sady B, Hudson M, Kim C, Apel K, Quail P H. 2004. Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science, 5692:1937-1941. |
| [33] |
Inoue K, Nishihama R, Kataoka H, Hosaka M, Manabe R, Nomoto M, Tada Y, Ishizaki K, Kohchi T. 2016. Phytochrome signaling is mediated by PHYTOCHROME INTERACTING FACTOR in the liverwort Marchantia polymorpha. Plant Cell, 28 (6):1406-1421.
doi: 10.1105/tpc.15.01063 URL |
| [34] |
Jia Y, Ding Y, Shi Y, Zhang X, Gong Z, Yang S. 2016. The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytologist, 212:345-353.
doi: 10.1111/nph.2016.212.issue-2 URL |
| [35] |
Jia Y, Kong X, Hu K, Cao M, Liu J, Ma C, Guo S, Xianzheng Y, Zhao S, Robert H S, Li C, Tian H, Ding Z. 2020. PIFs coordinate shade avoidance by inhibiting auxin repressor ARF18 and metabolic regulator QQS. The New Phytologist, 228 (2):609-621.
doi: 10.1111/nph.v228.2 URL |
| [36] |
Jiang B, Shi Y, Peng Y, Jia Y, Yan Y, Dong X, Li H, Dong J, Li J, Gong Z, Thomashow M F, Yang S. 2020. Cold-induced CBF-PIF 3 interaction enhances freezing tolerance by stabilizing the phyB thermosensor in Arabidopsis. Molecular Plant, 13 (6):894-906.
doi: 10.1016/j.molp.2020.04.006 URL |
| [37] | Jiang B, Shi Y, Zhang X, Xin X, Qi L, Guo H, Li J, Yang S. 2017. PIF3 is a negative regulator of the CBF pathway and freezing tolerance in Arabidopsis.. Proceedings of the National Academy of Sciences of the United States of America, 114 (32):E6695-E6702. |
| [38] |
Khanna R, Huq E, Kikis E A, Al-Sady B, Lanzatella C, Quail P H. 2004. A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific Basic Helix-Loop-Helix transcription factors. Plant Cell, 16 (11):3033-3044.
doi: 10.1105/tpc.104.025643 URL |
| [39] |
Kudo M, Kidokoro S, Yoshida T, Mizoi J, Todaka D, Fernie A R, Shinozaki K, Yamaguchi-Shinozaki K. 2017. Double overexpression of DREB and PIF transcription factors improves drought stress tolerance and cell elongation in transgenic plants. Plant Biotechnology Journal, 15 (4):458-471.
doi: 10.1111/pbi.2017.15.issue-4 URL |
| [40] |
Kumar I, Swaminathan K, Hudson K, Hudson M E. 2016. Evolutionary divergence of phytochrome protein function in Zea mays PIF3 signaling. Journal of Experimental Botany, 67 (14):4231-4240.
doi: 10.1093/jxb/erw217 URL |
| [41] |
Kumar S V, Lucyshyn D, Jaeger K E, Alos E, Alvey E, Harberd N P, Wigge P A. 2012. Transcription factor PIF4 controls the thermosensory activation of flowering. Nature, 484:242-245.
doi: 10.1038/nature10928 URL |
| [42] |
Kunihiro A, Yamashino T, Nakamichi N, Niwa Y, Nakanishi H, Mizuno T. 2011. PHYTOCHROME-INTERACTING FACTOR 4 and 5(PIF4 and PIF5)activate the homeobox ATHB2 and auxin-inducible IAA29 genes in the coincidence mechanism underlying photoperiodic control of plant growth of Arabidopsis thaliana. Plant and Cell Physiology, 52 (8):1315-1329.
doi: 10.1093/pcp/pcr076 URL |
| [43] |
Lau O S, Song Z, Zhou Z, Davies K A, Chang J, Yang X, Wang S, Lucyshyn D, Tay I H Z, Wigge P A, Bergmann D C. 2018. Direct control of SPEECHLESS by PIF 4 in the High-temperature response of stomatal development. Current Biology, 28 (8):1273-1280.
doi: 10.1016/j.cub.2018.02.054 |
| [44] | Lee C M, Thomashow M F. 2012. Photoperiodic regulation of the C-repeat binding factor(CBF)cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America, 109 (37):15054-15059. |
| [45] |
Lee K P, Piskurewicz U, Tureckova V, Carat S, Chappuis R, Strnad M, Fankhauser C, Lopez-Molina L. 2012. Spatially and genetically distinct control of seed germination by phytochromes A and B. Genes & Development, 26 (17):1984-1996.
doi: 10.1101/gad.194266.112 URL |
| [46] |
Lee N, Choi G. 2017. Phytochrome-interacting factor from Arabidopsis to liverwort. Current Opinion in Plant Biology, 35:54-60.
doi: 10.1016/j.pbi.2016.11.004 URL |
| [47] |
Leivar P, Monte E. 2014. PIFs:systems integrators in plant development. Plant Cell, 26 (1):56-78.
doi: 10.1105/tpc.113.120857 URL |
| [48] |
Leivar P, Quail P H. 2011. PIFs:pivotal components in a cellular signaling hub. Trends in Plant Science, 16 (1):19-28.
doi: 10.1016/j.tplants.2010.08.003 URL pmid: 20833098 |
| [49] |
Li K, Yu R, Fan L M, Wei N, Chen H, Deng X W. 2016. DELLA-mediated PIF degradation contributes to coordination of light and gibberellin signalling in Arabidopsis. Nature Communications, 7:11868.
doi: 10.1038/ncomms11868 URL |
| [50] | Li Lu, Wang Xiang, Wang Zhuang-yuan, Cao Yun, Hu Ge, Wang Liu-yi, Yin Jun. 2019. Cloning and expression analysis of phytochrome interacting factor TaPIF4 in wheat. Acta Agriculturae Boreali-Sinica, 34:74-82. (in Chinese) |
| 李璐, 王翔, 王状元, 曹云, 胡格, 王留壹, 尹钧. 2019. 小麦光敏色素互作因子TaPIF4基因的克隆与表达分析. 华北农学报, 34:78-82. | |
| [51] | Li Rui-ling, Ye Yun-tian, Jiang Lei-yu, Ye Yu-yun, Xiao Jie, Li Ya-li, Li Xin, Tang Hao-ru. 2018. Cloning and expression analysis of PIF3 gene in Fragaria × ananassa. Molecular Plant Breeding, 16 (19):6220-6227. (in Chinese) |
| 李瑞玲, 叶云天, 江雷雨, 叶宇芸, 肖婕, 李亚丽, 李欣, 汤浩茹. 2018. 草莓PIF3基因的克隆及表达分析. 分子植物育种, 16 (19):38-45. | |
| [52] | Ling J J, Li J, Zhu D, Deng X W. 2017. Noncanonical role of Arabidopsis COP1/SPA complex in repressing BIN2-mediated PIF 3 phosphorylation and degradation in darkness. Proceedings of the National Academy of Sciences of the United States of America, 114 (13):3539-3544. |
| [53] |
Lockhart J. 2013. Frenemies:antagonistic bHLH/bZIP transcription factors integrate light and reactive oxygen species signaling in Arabidopsis. Plant Cell, 25 (5):1483.
doi: 10.1105/tpc.113.250510 URL |
| [54] |
Lorenzo C D, Alonso Iserte J, Sanchez Lamas M, Sofia Antonietti M, Garcia Gagliardi P, Hernando C E, Dezar C A A, Vazquez M, Casal J J, Yanovsky M J, Cerdan P D. 2019. Shade delays flowering in Medicago sativa. Plant Journal, 99 (1):7-22.
doi: 10.1111/tpj.2019.99.issue-1 URL |
| [55] | Ma D, Li X, Guo Y, Chu J, Fang S, Yan C, Noel J P, Liu H. 2016. Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proceedings of the National Academy of Sciences of the United States of America, 113 (1):224-229. |
| [56] | Miao Yanxiu, Chen Qingyun, Qu Mei, Gao Lihong, Hou Leiping, Li Bin. 2019. Effects of red and blue lights on growth,photosynthetic characteristics and yield of cucumber plants. Acta Horticulturae Sinica, 46 (7):1388-1398. (in Chinese) |
| 苗妍秀, 陈青云, 曲梅, 高丽红, 侯雷平, 李斌. 2019. 黄瓜红蓝光质育苗对其定植后生长、光合特性以及产量的影响. 园艺学报, 46 (7):1388-1398. | |
| [57] |
Ni M, Tepperman J M, Quail P H. 1998. PIF3,a phytochrome-interacting factor necessary for normal photoinduced signal transduction,is a novel Basic Helix-Loop-Helix protein. Cell, 95 (5):657-667.
pmid: 9845368 |
| [58] |
Ni W, Xu S L, Gonzalez-Grandio E, Chalkley R J, Huhmer A F R, Burlingame A L, Wang Z Y, Quail P H. 2017. PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3. Nature Communications, 8:15236.
doi: 10.1038/ncomms15236 URL |
| [59] |
Ni W, Xu S L, Tepperman J M, Stanley D J, Maltby D A, Gross J D, Burlingame A L, Wang Z Y, Quail P H. 2014. A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science, 344 (6188):1160-1164.
doi: 10.1126/science.1250778 URL |
| [60] |
Nieto C, Lopez-Salmeron V, Daviere J M, Prat S. 2015. ELF3-PIF 4 Interaction regulates plant growth independently of the evening complex. Current Biology, 25 (2):187-193.
doi: 10.1016/j.cub.2014.10.070 URL |
| [61] |
Noir S, Boemer M, Takahashi N, Ishida T, Tsui T L, Balbi V, Shanahan H, Sugimoto K, Devoto A. 2013. Jasmonate controls leaf growth by repressing cell proliferation and the onset of endoreduplication while maintaining a potential stand-by mode. Plant Physiology, 161 (4):1930-1951.
doi: 10.1104/pp.113.214908 URL |
| [62] |
Nusinow D A, Helfer A, Hamilton E E, King J J, Imaizumi T, Schultz T F, Farré E M, Kay S A. 2011. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature, 475 (7356):398-402.
doi: 10.1038/nature10182 pmid: 21753751 |
| [63] |
Oh E, Kim J, Park E, Kim J-I, Kang C, Choi G. 2004. PIL5,a phytochrome-interacting basic helix-loop-helix protein,is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell, 16 (11):3045-3058.
doi: 10.1105/tpc.104.025163 URL |
| [64] |
Oh E, Yamaguchi S, Hu J, Yusuke J, Jung B, Paik I, Lee H S, Sun T P, Kamiya Y, Choi G. 2007. PIL5,a phytochrome-interacting bHLH protein,regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell, 19 (4):1192-1208.
doi: 10.1105/tpc.107.050153 URL |
| [65] |
Oh E, Zhu J Y, Wang Z Y. 2012. Interaction between BZR1 and PIF 4 integrates brassinosteroid and environmental responses. Nature Cell Biology, 14 (8):802-809.
doi: 10.1038/ncb2545 URL |
| [66] |
Pedmale U V, Huang S S C, Zander M, Cole B J, Hetzel J, Ljung K, Reis P A B, Sridevi P, Nito K, Nery J R, Ecker J R, Chory J. 2016. Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell, 164 (1-2):233-245.
doi: 10.1016/j.cell.2015.12.030 URL |
| [67] |
Peng M, Li Z, Zhou N, Ma M, Jiang Y, Dong A, Shen W H, Li L. 2018. Linking PHYTOCHROME-INTERACTING FACTOR to histone modification in plant shade avoidance. Plant Physiology, 176 (2):1341-1351.
doi: 10.1104/pp.17.01189 URL |
| [68] |
Pieterse Corné M J, Zamioudis C, Berendsen R L, Weller D M, van Wees S C M, Bakker P A H M. 2014. Induced systemic resistance by beneficial microbes. Annual Review of Phytopathology, 52:347-375.
doi: 10.1146/annurev-phyto-082712-102340 URL pmid: 24906124 |
| [69] |
Possart A, Xu T F, Paik I, Hanke S, Keim S, Hermann H M, Wolf L, Hiss M, Becker C, Huq E, Rensing S A, Hiltbrunner A. 2017. Characterization of phytochrome interacting factors from the moss Physcomitrella patens illustrates conservation of phytochrome signaling modules in land plants. Plant Cell, 29 (2):310-330.
doi: 10.1105/tpc.16.00388 URL |
| [70] | Pucciariello O, Legris M, Costigliolo R C, Iglesias M J, Hernando C E, Dezar C, Vazquez M, Yanovsky M J, Finlayson S A, Prat S, Prat S, Casal J J. 2018. Rewiring of auxin signaling under persistent shade. Proceedings of the National Academy of Sciences of the United States of America, 115 (21):5612-5617. |
| [71] | Qiao Yu. 2012. Molecular cloning and functional characterization of an apple Phytochrome-Interacting Factor gene MdPIF1[M. D. Dissertation]. Taian:Shandong Agricultural University. (in Chinese) |
| 谯愚. 2012. 苹果光敏色素互作因子基因 MdPIF1的克隆和功能鉴定[硕士论文]. 泰安:山东农业大学. | |
| [72] |
Qiu J R, Xiang X Y, Wang J T, Xu W X, Chen J, Xiao Y, Jiang C Z, Huang Z. 2020. MfPIF1 of resurrection plant Myrothamnus flabellifolia plays a positive regulatory role in responding to drought and salinity stresses in Arabidopsis. International Journal of Molecular Sciences, 21 (8):3011.
doi: 10.3390/ijms21083011 URL |
| [73] |
Qiu Y, Li M, Kim R J-A, Moore C M, Chen M. 2019. Daytime temperature is sensed by phytochrome B in Arabidopsis through a transcriptional activator HEMERA. Nature Communications, 10:140.
doi: 10.1038/s41467-018-08059-z URL |
| [74] | Quail P H. 2000. Phytochrome-interacting factors. Seminars in Cell & Developmental Biology, 11 (6):457-466. |
| [75] | Ren Xiao-yun. 2017. Cloning and expression of ZmPIFs and study on the drought resistant function of AtPIFs[M. D. Dissertation]. Yangzhou:Yangzhou University. (in Chinese) |
| 任小芸. 2017. ZmPIFs基因的克隆、表达及 AtPIFs基因的抗旱功能研究[硕士论文]. 扬州:扬州大学. | |
| [76] | Ren Xiao-yun, Wu Mei-qin, Chen Jian-min, Zhang Dong-ping, Gao Yong. 2016. The molecular mechanisms of phytochrome interacting factors (PIFs)in phytohormone signaling transduction. Plant Physiology Communications, 52 (10):1466-1473. (in Chinese) |
| 任小芸, 吴美琴, 陈建民, 张冬平, 高勇. 2016. 光敏色素作用因子PIFs参与植物激素信号转导的分子机制. 植物生理学报, 52 (10):1466-1473. | |
| [77] |
Ren Y R, Zhao Q, Zhao X Y, Hao Y J, You C X. 2016. Expression analysis of the MdCIbHLH1gene in apple flower buds and seeds in the process of dormancy. Horticultural Plant Journal, 2 (2):61-66.
doi: 10.1016/j.hpj.2016.06.007 URL |
| [78] |
Rosado D, Gramegna G, Cruz A, Lira B S, Freschi L, de Setta N, Rossi M. 2016. Phytochrome interacting factors(PIFs)in Solanum lycopersicum:diversity,evolutionary history and expression profiling during different developmental processes. PLoS ONE, 11 (11):e0165929.
doi: 10.1371/journal.pone.0165929 URL |
| [79] |
Seo M, Nambara E, Choi G, Yamaguchi S. 2009. Interaction of light and hormone signals in germinating seeds. Plant Molecular Biology, 69 (4):463-472.
doi: 10.1007/s11103-008-9429-y URL |
| [80] |
Sewelam N, Kazan K, Schenk P M. 2016. Global plant stress signaling:reactive oxygen species at the cross-road. Frontiers in Plant Science, 7:187.
doi: 10.3389/fpls.2016.00187 pmid: 26941757 |
| [81] |
Sharma A, Sharma B, Hayes S, Kerner K, Hoecker U, Jenkins G I, Franklin K A. 2019. UVR8 disrupts stabilisation of PIF 5 by COP1 to inhibit plant stem elongation in sunlight. Nature Communications, 10 (1):4417.
doi: 10.1038/s41467-019-12369-1 URL |
| [82] |
Shen H, Zhu L, Castillon A, Majee M, Downie B, Huq E. 2008. Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell, 20 (6):1586-1602.
doi: 10.1105/tpc.108.060020 pmid: 18539749 |
| [83] | Shuai Hai-wei, Meng Yong-jie, Chen Feng, Zhou Wen-guan, Luo Xiao-feng, Yang Wen-yu, Shu Kai. 2018. Phytohormone-mediated plant shade responses. Journal of Integrative, 53:139-148. (in Chinese) |
| 帅海威, 孟永杰, 陈锋, 周文冠, 罗晓峰, 杨文钰, 舒凯. 2018. 植物荫蔽胁迫的激素信号响应. 植物学报, 53:139-148. | |
| [84] | Song Yang, Zhang Yan-min, Wang Chuan-zeng, Liu Mei-yan, Liu Jin, Wang Yan-ling, Chen Xue-sen. 2012. Clone and expression analyzing of phytochrome-interacting factor gene PIF of spur type apple. Acta Horticulturae Sinica, 39 (4):743-748. (in Chinese) |
| 宋杨, 张艳敏, 王传增, 刘美艳, 刘金, 王延玲, 陈学森. 2012. 苹果光敏色素作用因子基因PIF的克隆和分析. 园艺学报, 39 (4):743-748. | |
| [85] |
Sun T P. 2010. Gibberellin-GID1-DELLA:a pivotal regulatory module for plant growth and development. Plant Physiology, 154 (2):567-570.
doi: 10.1104/pp.110.161554 URL |
| [86] |
Sun W J, Han H Y, Deng L, Sun C L, Xu Y R, Lin L H, Ren P R, Zhao J H, Zhai Q Z, Li C Y. 2020. Mediator subunit MED 25 physically interacts with PHYTOCHROME INTERACTING FACTOR 4PIF4 to regulate shade-induced hypocotyl elongation in tomato. Plant Physiology, 184 (3):1549-1562.
doi: 10.1104/pp.20.00587 URL |
| [87] |
Tavridou E, Pireyre M, Ulm R. 2020. Degradation of the transcription factors PIF4 and PIF 5 under UV-B promotes UVR8-mediated inhibition of hypocotyl growth in Arabidopsis. Plant Journal, 101 (3):507-517.
doi: 10.1111/tpj.14556 |
| [88] |
Thines B C, Youn Y, Duarte M I, Harmon F G. 2014. The time of day effects of warm temperature on flowering time involve PIF4 and PIF5. Journal of Experimental Botany, 65 (4):1141-1151.
doi: 10.1093/jxb/ert487 URL pmid: 24574484 |
| [89] | Wang Feng. 2017. Roles and mechanisms of phyA-,HY5- and PIF4- mediated light quality-regulated cold tolerance in tomato[Ph. D. Dissertation]. Hangzhou:Zhejiang University. (in Chinese) |
| 王峰. 2017. PhyA、HY5和PIF4在光质调控番茄低温抗性中的机制研究[博士论文]. 杭州:浙江大学. | |
| [90] | Wang Hong. 2009. Effects of light quality on photosynthetic efficiency and the resistance to Sphaerotheca fuliginea in cucumber plants[Ph. D. Dissertation]. Hangzhou:Zhejiang University. (in Chinese) |
| 王虹. 2009. 光质对黄瓜幼苗光合效率和白粉病抗性的调控机理[博士论文]. 杭州:浙江大学. | |
| [91] | Wang Shuo, Ding Lan, Liu Jian-xiang, Han Jia-jia. 2018. PIF4-regulated thermo-responsive genes in Arabidopsis. Biotechnology Bulletin, 34:57-65. (in Chinese) |
| 汪硕, 丁岚, 刘建祥, 韩佳嘉. 2018. 拟南芥热形态建成中PIF4下游基因研究. 生物技术通报, 34:57-65. | |
| [92] | Wang Xiaodi, Ji Xiaohao, Zheng Xiaocui, Wang Yingying, Song Yang, Liu Fengzhi. 2019. Cloning and functional analysis of a cold treatment response factor gene PdCIbHLH in peach. Acta Horticulturae Sinica, 46 (3):444-452. (in Chinese) |
| 王孝娣, 冀晓昊, 郑晓翠, 王莹莹, 宋杨, 刘凤之. 2019. 冷处理响应基因 PdCIbHLH的克隆和功能鉴定. 园艺学报, 46 (3):444-452. | |
| [93] |
Windram O, Madhou P, McHattie S, Hill C, Hickman R, Cooke E, Jenkins D J, Penfold C A, Baxter L, Breeze E, Kiddle S J, Rhodes J, Atwell S, Kliebenstein D J, Kim Y S, Stegle O, Borgwardt K, Zhang C, Tabrett A, Legaie R, Moore J, Finkenstadt B, Wild D L, Mead A, Rand D, Beynon J, Ott S, Buchanan-Wollaston V, Denby K J. 2012. Arabidopsis defense against Botrytis cinerea:chronology and regulation deciphered by high-resolution temporal transcriptomic analysis. Plant Cell, 24 (9):3530-3557.
doi: 10.1105/tpc.112.102046 URL |
| [94] |
Xiang S Y, Wu S G, Zhang H Y, Mou M H, Chen Y L, Li D B, Wang H P, Chen L G, Yu D Q. 2020. The PIFs redundantly control plant defense response against Botrytis cinerea in Arabidopsis. Plants, 9 (9):1246.
doi: 10.3390/plants9091246 URL |
| [95] | Xu X, Kathare P K, Vinh Ngoc P, Bu Q, Nguyen A, Huq E. 2017. Reciprocal proteasome-mediated degradation of PIFs and HFR1 underlies photomorphogenic development in Arabidopsis. Development, 144 (10):1831-1840. |
| [96] |
Xu X, Paik I, Zhu L, Huq E. 2015. Illuminating progress in phytochrome-mediated signaling pathways. Trends in Plant Science, 20 (10):641-650.
doi: 10.1016/j.tplants.2015.06.010 URL |
| [97] | Xu Xiang-dong, Ren Yi-qiu, Zhang Li, Li Yu, Wang Li-juan, Lu Meng-zhu. 2018. Analysis of expression pattern of PIF family members in populus. Forest Research, 31:19-25. (in Chinese) |
| 徐向东, 任逸秋, 张利, 李煜, 王丽娟, 卢孟柱. 2018. 杨树PIF基因家族成员表达模式研究. 林业科学研究, 31:19-25. | |
| [98] |
Yamaguchi-Shinozaki K, Shinozaki K. 2006. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annual Review of Plant Biology, 57:781-803.
pmid: 16669782 |
| [99] | Yan Y, Li C, Dong X, Li H, Zhang D, Zhou Y, Jiang B, Peng J, Qin X, Cheng J, Wang X, Song P, Qi L, Zheng Y, Li B, Terzaghi W, Yang S, Guo Y, Li J. 2020. MYB 30 is a key negative regulator of Arabidopsis photomorphogenic development that promotes PIF4 and PIF5 protein accumulation in the light. Plant Cell,tpc.00645. |
| [100] | Yang D L, Yao J, Mei C S, Tong X H, Zeng L J, Li Q, Xiao L T, Sun T P, Li J, Deng X W, Lee C M, Thomashow M F, Yang Y, He Z, He S Y. 2012. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proceedings of the National Academy of Sciences of the United States of America, 109 (19):E1192-E1200. |
| [101] | Yang Jian-fei, Wang Yu, Yang Lin, Li Yu-hua. 2014. Phytochrome-interacting factors integrate multiple signals to control plant growth and development. Plant Physiology Communications, 50 (8):1109-1118. |
| 杨剑飞, 王宇, 杨琳, 李玉花. 2014. 光敏色素互作因子PIFs是整合多种信号调控植物生长发育的核心元件. 植物生理学报, 50 (8):1109-1118. | |
| [102] |
Zhang B, Holmlund M, Lorrain S, Norberg M, Bako L, Fankhauser C, Nilsson O. 2017. BLADE-ON-PETIOLE proteins act in an E3 ubiquitin ligase complex to regulate PHYTOCHROME INTERACTING FACTOR 4 abundance. eLife, 6:e26759.
doi: 10.7554/eLife.26759 URL |
| [103] |
Zhang K K, Zheng T, Zhu X D, Jiu S T, Liu Z J, Guan L, Jia H F, Fang J G. 2018. Genome-wide identification of PIFs in grapes( Vitis vinifera L.)and their transcriptional analysis under lighting/shading conditions. Genes, 9 (9):451.
doi: 10.3390/genes9090451 URL |
| [104] | Zhang Meng-jiao, Zhu Guang, Tao Hai-xia, Chen Jian-min, Gao Yong. 2019. Research advances of PIFs in plant growth and development. Chinese Journal of Cell Biology, 41 (8):1633-1639. (in Chinese) |
| 张梦娇, 朱光, 陶海霞, 陈建民, 高勇. 2019. 光敏色素作用因子PIFs在植物生长发育中的研究进展. 中国细胞生物学学报, 41 (8):1633-1639. | |
| [105] |
Zhang R, Yang C, Jiang Y, Li L. 2019. A PIF7-CONSTANS-centered molecular regulatory network underlying shade-accelerated flowering. Molecular Plant, 12 (12):1587-1597.
doi: 10.1016/j.molp.2019.09.007 URL |
| [106] |
Zhang Y, Mayba O, Pfeiffer A, Shi H, Tepperman J M, Speed T P, Quail P H. 2013. A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genetics, 9 (1):e1003244.
doi: 10.1371/journal.pgen.1003244 URL |
| [107] |
Zhao C, Zhang Z, Xie S, Si T, Li Y, Zhu J K. 2016. Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiology, 171 (4):2744-2759.
doi: 10.1104/pp.16.00533 URL |
| [108] | Zheng Lan-lan, Li Chen, Zhang Jing-xuan, Lan Hong-meng, Zhou Meng, Zhang Yong-hong. 2017. Hypocotyl elongation of Arabidopsis under high temperature condition through PIF 4 gene control regulated by HY5. Hubei Agricultural Sciences, 56 (18):3554-3558. (in Chinese) |
| 郑兰兰, 李琛, 张璟璇, 兰红梦, 周蒙, 张勇洪. 2017. HY5通过PIF4基因控制高温诱导的拟南芥下胚轴伸长反应. 湖北农业科学, 56 (18):3554-3558. | |
| [109] | Zhou Feng. 2015. The plant defense response and its light signaling regulation pathways. Northern Horticulture,(17):179-182. (in Chinese) |
| 周峰. 2015. 植物防御反应及其光信号调控途径. 北方园艺,(17):179-182. | |
| [110] |
Zhou L J, Mao K, Qiao Y, Jiang H, Li Y Y, Hao Y J. 2017. Functional identification of MdPIF1 as a phytochrome interacting factor in apple. Plant Physiology and Biochemistry, 119:178-188.
doi: 10.1016/j.plaphy.2017.08.027 URL |
| [111] |
Zhou Y, Xun Q, Zhang D, Lü M, Ou Y, Li J. 2019. TCP transcription factors associate with PHYTOCHROME INTERACTING FACTOR 4 and CRYPTOCHROME 1 to regulate thermomorphogenesis in Arabidopsis thaliana. iScience, 15:600-610.
doi: S2589-0042(19)30099-9 pmid: 31078553 |
| [112] | Zhou Yu. 2018. TCP transcription factors regulate shade avoidance via promoting the activity of PIFs and the expression of auxin biosynthetic genes[Ph. D. Dissertation]. Lanzhou:Lanzhou University. (in Chinese) |
| 周瑜. 2018. TCP转录因子通过促进PIFs活性和生长素合成基因的表达调控避荫反应[博士论文]. 兰州:兰州大学. | |
| [113] |
Zhu L, Bu Q, Xu X, Paik I, Huang X, Hoecker U, Deng X W, Huq E. 2015. CUL 4 forms an E3 ligase with COP1 and SPA to promote light-induced degradation of PIF1. Nature Communications, 6:7245.
doi: 10.1038/ncomms8245 URL |
| [1] | 徐小萍, 曹清影, 蔡柔荻, 官庆栩, 张梓浩, 陈裕坤, 徐涵, 林玉玲, 赖钟雄. 龙眼miR408与DlLAC12克隆及其在球形胚发生和非生物胁迫下的表达分析[J]. 园艺学报, 2022, 49(9): 1866-1882. |
| [2] | 贾鑫, 曾臻, 陈月, 冯慧, 吕英民, 赵世伟. 月季‘月月粉’RcDREB2A的克隆与表达分析[J]. 园艺学报, 2022, 49(9): 1945-1956. |
| [3] | 马维峰, 李艳梅, 马宗桓, 陈佰鸿, 毛娟. 苹果POD家族基因的鉴定与MdPOD15的功能分析[J]. 园艺学报, 2022, 49(6): 1181-1199. |
| [4] | 周至铭, 杨佳宝, 张程, 曾令露, 孟晚秋, 孙黎. 向日葵LACS家族鉴定及响应非生物胁迫表达分析[J]. 园艺学报, 2022, 49(2): 352-364. |
| [5] | 王云, 张镇武, 孙逊, 张绍铃. 植物自噬与病原菌互作研究进展[J]. 园艺学报, 2022, 49(10): 2205-2222. |
| [6] | 谢思艺, 周承哲, 朱晨, 詹冬梅, 陈兰, 吴祖春, 赖钟雄, 郭玉琼. 茶树CsTIFY家族全基因组鉴定及非生物胁迫和激素处理中主要基因表达分析[J]. 园艺学报, 2022, 49(1): 100-116. |
| [7] | 梁志乐, 汪宽鸿, 杨静, 祝彪, 朱祝军. 硫代葡萄糖苷在十字花科植物应对非生物胁迫中的作用[J]. 园艺学报, 2022, 49(1): 200-220. |
| [8] | 杨天宸, 陈晓童, 吕可, 张荻. 百子莲脱水素基因ApSK3对逆境与激素信号的应答模式与调控机制[J]. 园艺学报, 2021, 48(8): 1565-1578. |
| [9] | 马俊杰, 宋丽娜, 李乐, 马晓春, 靳磊, 徐伟荣. 山葡萄VaCBL6参与非生物胁迫和ABA途径的响应[J]. 园艺学报, 2021, 48(6): 1079-1093. |
| [10] | 汪宽鸿, 祝彪, 朱祝军. GSH/GSSG在植物应对非生物胁迫中的作用综述[J]. 园艺学报, 2021, 48(4): 647-660. |
| [11] | 贾兵, 郭国凌, 王友煜, 叶振风, 刘莉, 刘普, 衡伟, 朱立武. ‘黄金梨’缺铁黄化叶片受GA3诱导复绿的机理研究[J]. 园艺学报, 2021, 48(2): 254-264. |
| [12] | 白露, 张志国, 张世杰, 黄东梅, 秦巧平. 萱草3种蔗糖转化酶基因的分离及对低温和渗透胁迫响应的分析[J]. 园艺学报, 2021, 48(2): 300-312. |
| [13] | 张婷婷, 李雨欣, 张德遥, 康宇乾, 王健, 宋希强, 周扬. 铁皮石斛蛋白磷酸酶PP2C家族基因鉴定及其表达分析[J]. 园艺学报, 2021, 48(12): 2458-2470. |
| [14] | 柯玉洁, 陈明堃, 马山虎, 欧悦, 王艺, 郑清冬, 刘仲健, 艾叶. 兰科植物MYB转录因子研究进展[J]. 园艺学报, 2021, 48(11): 2311-2320. |
| [15] | 李欣欣, 侯鸿敏, 徐吉花, 孙晓红, 张玉刚. 苹果MLP家族基因鉴定及非生物胁迫响应分析[J]. 园艺学报, 2021, 48(1): 1-14. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司