
园艺学报 ›› 2021, Vol. 48 ›› Issue (3): 553-565.doi: 10.16420/j.issn.0513-353x.2020-0465
收稿日期:2020-11-24
出版日期:2021-03-25
发布日期:2021-04-02
通讯作者:
李淑娴
E-mail:shuxianli@njfu.com.cn
基金资助:
CHEN Li, XUE Liangjiao, LI Shuxian( )
)
Received:2020-11-24
Online:2021-03-25
Published:2021-04-02
Contact:
LI Shuxian
E-mail:shuxianli@njfu.com.cn
摘要:
跳枝碧桃(Prunus persicaf. versicolor)在同一植株上可产生二色花和嵌合花,具有很高的观赏价值。以南京3个观赏桃集中分布区的57株跳枝碧桃为材料,利用简单重复序列(SSR)标记研究其遗传背景。通过PCR扩增,筛选出6对多态性引物,共计扩增出17个多态性条带,平均有效等位基因2.83个。SSR标记基因分型结果表明,供试材料可分为3种基因型,各采样地均具有2 ~ 3种基因型,同一植株的不同分枝基因型完全一致。基于基因分型结果,选取3种基因型跳枝碧桃各1株进行基因组重测序,结合公开发表的数据,进行位点变异分析,并构建系统发育树。结果显示3种基因型的跳枝碧桃聚在一个分支,其中B20、T3基因型与‘洒红桃’聚在一起,W5基因型相对独立。3种基因型的跳枝碧桃在起源上比较接近,但不同基因型之间存在一定的遗传差异。通过整合单核苷酸多态性(SNP)信息和花色表型开展全基因组关联分析(GWAS),鉴定出5个SNP与花色跳枝性状显著关联(P值 < 10 -100),均位于7号染色体。在显著SNP位点的邻近基因组区段发掘3个DICER-LIKE 2(DCL2)基因,揭示在桃树花色跳枝性状的形成中,依赖DCL2的表观遗传修饰途径可能发挥重要作用。
中图分类号:
陈丽, 薛良交, 李淑娴. 跳枝碧桃花色性状的全基因组关联分析[J]. 园艺学报, 2021, 48(3): 553-565.
CHEN Li, XUE Liangjiao, LI Shuxian. Genome-wide Association Study of Flower Color Trait in Prunus persicaf.versicolor[J]. Acta Horticulturae Sinica, 2021, 48(3): 553-565.

图1 跳枝碧桃的花色特征 A:同一枝条上开两种颜色的花;B:同一朵花上有两种颜色的花瓣。
Fig. 1 Variegated flowers of Purnus persica f. versicolor A shows flowers of two colors on the same branch,and B shows petals of two colors on the same flower.
图3 应用琼脂糖凝胶电泳方法检测桃花基因组DNA质量 Marker为DNA长度标记,其余泳道为3个采样地的代表样品,其中r和w分别表示红花和白花。下同。
Fig. 3 Examination of peach genomic DNA samples using agarose gel electrophoresis Maker indicates DNA size markers,the other lanes are presentative samples from three loci. r and w indicated red and white flowers,respectively. The same below.
| 引物编号 Primer ID | 重复单元 Repeat motif | 产物大小/bp Allele size range | 多态性条带数 Number of polymorphic alleles | 多态性频率/% Percentage of polymorphism | 参考文献 Reference |
|---|---|---|---|---|---|
| BPPCT014 | (AG)23 | 197 ~ 224 | 3 | 100.0 | Dirlewanger et al. |
| BPPCT017 | (GA)28 | 150 ~ 180 | 4 | 100.0 | Dirlewanger et al. |
| BPPCT025 | (GA)29 | 175 ~ 188 | 3 | 100.0 | Dirlewanger et al. |
| BPPCT038 | (GA)25 | 157 ~ 170 | 2 | 100.0 | Dirlewanger et al. |
| UDP97-401 | (GA)19 | 124 ~ 144 | 2 | 66.7 | Cipriani et al. 1999 |
| UDP98-409 | (AG)19 | 120 ~ 183 | 3 | 100.0 | Cipriani et al. 1999 |
表1 多态性引物的扩增情况统计
Table 1 Summary of amplification results using polymorphic primers
| 引物编号 Primer ID | 重复单元 Repeat motif | 产物大小/bp Allele size range | 多态性条带数 Number of polymorphic alleles | 多态性频率/% Percentage of polymorphism | 参考文献 Reference |
|---|---|---|---|---|---|
| BPPCT014 | (AG)23 | 197 ~ 224 | 3 | 100.0 | Dirlewanger et al. |
| BPPCT017 | (GA)28 | 150 ~ 180 | 4 | 100.0 | Dirlewanger et al. |
| BPPCT025 | (GA)29 | 175 ~ 188 | 3 | 100.0 | Dirlewanger et al. |
| BPPCT038 | (GA)25 | 157 ~ 170 | 2 | 100.0 | Dirlewanger et al. |
| UDP97-401 | (GA)19 | 124 ~ 144 | 2 | 66.7 | Cipriani et al. 1999 |
| UDP98-409 | (AG)19 | 120 ~ 183 | 3 | 100.0 | Cipriani et al. 1999 |
| 基因型 Genotype | 植株编号 Plant accession | 枝条 Branch | 花色 Color |
|---|---|---|---|
| Ⅰ | W1 ~ W8,B1 | 每株1 ~ 3个枝条 1-3 branches per plant | 每个枝条上红、白色花 Red and white flowers on each branch |
| Ⅱ | W9,B2 ~ B19,B21,B23,B25,T1 ~ T18,T20 ~ T22 | 每株1 ~ 3个枝条 1-3 branches per plant | 每个枝条上红、白色花 Red and white flowers on each branch |
| Ⅲ | W10,B20,B22,B24,T19 | 每株1 ~ 3个枝条 1-3 branches per plant | 每个枝条上红、白色花 Red and white flowers on each branch |
表2 供试跳枝碧桃材料的基因型
Table 2 Genotypes of tested peach samples
| 基因型 Genotype | 植株编号 Plant accession | 枝条 Branch | 花色 Color |
|---|---|---|---|
| Ⅰ | W1 ~ W8,B1 | 每株1 ~ 3个枝条 1-3 branches per plant | 每个枝条上红、白色花 Red and white flowers on each branch |
| Ⅱ | W9,B2 ~ B19,B21,B23,B25,T1 ~ T18,T20 ~ T22 | 每株1 ~ 3个枝条 1-3 branches per plant | 每个枝条上红、白色花 Red and white flowers on each branch |
| Ⅲ | W10,B20,B22,B24,T19 | 每株1 ~ 3个枝条 1-3 branches per plant | 每个枝条上红、白色花 Red and white flowers on each branch |
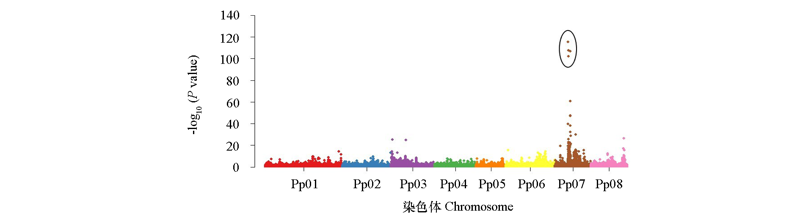
图6 跳枝碧桃花色跳枝性状的全基因组关联分析 椭圆圈内为与花色跳枝性状显著相关的SNP位点。
Fig. 6 Genome-wide association study of variegated flower color trait in Purnus persica f. versicolor In the ellipse,there were the SNP sites that are significantly associated with the traits of flower color.
| 基因编号 | 注释信息 | 基因编号 | 注释信息 |
|---|---|---|---|
| Gene ID | Annotation | Gene ID | Annotation |
| Prupe.7G044200 | 富含亮氨酸重复的蛋白激酶 | Prupe.7G047500 | 类转氨酶,植物移动域家族蛋白 |
| Leucine-rich repeat protein kinase | Aminotransferase-like,plant mobile domain family protein | ||
| Prupe.7G044300 | 富含亮氨酸重复的蛋白激酶 | Prupe.7G047600 | 钙调素结合蛋白 |
| Leucine-rich repeat protein kinase | Calmodulin binding | ||
| Prupe.7G044400 | 核酸内切酶Z4 tRNAse Z4 | Prupe.7G047700 | 类Dicer蛋白2 Dicer-like 2 |
| Prupe.7G044500 | 春化相关蛋白1 | Prupe.7G047900 | 类Dicer蛋白2 |
| Related to vernalization 1 | Dicer-like 2 | ||
| Prupe.7G044700 | 富含亮氨酸重复的蛋白激酶 | Prupe.7G048000 | 类Dicer蛋白2 |
| Leucine-rich repeat protein kinase | Dicer-like 2 | ||
| Prupe.7G044800 | 类AP2/B3转录因子 | Prupe.7G048100 | 富含羟脯氨酸的糖蛋白 |
| AP2/B3-like transcriptional factor | Hydroxyproline-rich glycoprotein | ||
| Prupe.7G044900 | 类HAL3蛋白A | Prupe.7G048300 | 类GDSL脂肪酶/酰水解酶 |
| HAL3-like protein A | GDSL-like lipase/acylhydrolase | ||
| Prupe.7G045000 | 甲硫氨酸依赖性甲基转移酶 | Prupe.7G048400 | 类SAUR生长素响应蛋白 |
| SAM-dependent methyltransferases | SAUR-like auxin-responsive | ||
| Prupe.7G045400 | 吡咯啉-5-羧酸盐还原酶 | Prupe.7G048500 | 类SAUR生长素响应蛋白 |
| P5C reductase | SAUR-like auxin-responsive | ||
| Prupe.7G045500 | 吡咯啉-5-羧酸盐还原酶 | Prupe.7G048600 | 类SAUR生长素响应蛋白 |
| P5C reductase | SAUR-like auxin-responsive | ||
| Prupe.7G045600 | 未知蛋白结构247 DUF247 | Prupe.7G055900 | AGL18蛋白AGAMOUS-like 18 |
| Prupe.7G045700 | 未知蛋白结构247 | Prupe.7G056000 | 未知蛋白结构617 |
| DUF247 | Protein of unknown function,DUF617 | ||
| Prupe.7G045800 | 未知蛋白结构247 | Prupe.7G056100 | 含P环的核苷三磷酸水解酶 |
| DUF247 | P-loop containing nucleoside triphosphate hydrolases | ||
| Prupe.7G046800 | 含ankyrin重复/BTB/ POZ结构域蛋白 | Prupe.7G056200 | ABC-2型转运蛋白 |
| Ankyrin repeat family protein/BTB/POZ domain- containing protein | ABC-2 type transporter protein | ||
| Prupe.7G047000 | 甲硫氨酸依赖的甲基转移酶 | Prupe.7G056300 | 含P环的核苷三磷酸水解酶 |
| SAM-dependent methyltransferases | P-loop containing nucleoside triphosphate hydrolases | ||
| Prupe.7G047100 | PPR蛋白 | Prupe.7G056400 | ABC-2型转运蛋白 |
| Pentatricopeptide repeat protein | ABC-2 type transporter | ||
| Prupe.7G047200 | 逆转录酶 | Prupe.7G056500 | NADPH依赖的硫氧还蛋白还原酶C |
| Reverse transcriptase | NADPH-dependent thioredoxin reductase C | ||
| Prupe.7G047300 | EF-hand钙结合蛋白 | Prupe.7G056700 | 热激转录因子B3 |
| Calcium-binding EF-hand protein | Heat shock transcription factor B3 | ||
| Prupe.7G047400 | 类Patatin磷脂酶家族蛋白 | Prupe.7G056900 | SEC14细胞溶质因子/磷酸甘油酯转移酶 |
| Patatin-like phospholipase family protein | SEC14 cytosolic factor/phosphoglyceride transfer |
表3 显著关联SNP位点附近的基因及其注释信息
Table 3 IDs and annotations of genes adjacent to significant associated SNPs
| 基因编号 | 注释信息 | 基因编号 | 注释信息 |
|---|---|---|---|
| Gene ID | Annotation | Gene ID | Annotation |
| Prupe.7G044200 | 富含亮氨酸重复的蛋白激酶 | Prupe.7G047500 | 类转氨酶,植物移动域家族蛋白 |
| Leucine-rich repeat protein kinase | Aminotransferase-like,plant mobile domain family protein | ||
| Prupe.7G044300 | 富含亮氨酸重复的蛋白激酶 | Prupe.7G047600 | 钙调素结合蛋白 |
| Leucine-rich repeat protein kinase | Calmodulin binding | ||
| Prupe.7G044400 | 核酸内切酶Z4 tRNAse Z4 | Prupe.7G047700 | 类Dicer蛋白2 Dicer-like 2 |
| Prupe.7G044500 | 春化相关蛋白1 | Prupe.7G047900 | 类Dicer蛋白2 |
| Related to vernalization 1 | Dicer-like 2 | ||
| Prupe.7G044700 | 富含亮氨酸重复的蛋白激酶 | Prupe.7G048000 | 类Dicer蛋白2 |
| Leucine-rich repeat protein kinase | Dicer-like 2 | ||
| Prupe.7G044800 | 类AP2/B3转录因子 | Prupe.7G048100 | 富含羟脯氨酸的糖蛋白 |
| AP2/B3-like transcriptional factor | Hydroxyproline-rich glycoprotein | ||
| Prupe.7G044900 | 类HAL3蛋白A | Prupe.7G048300 | 类GDSL脂肪酶/酰水解酶 |
| HAL3-like protein A | GDSL-like lipase/acylhydrolase | ||
| Prupe.7G045000 | 甲硫氨酸依赖性甲基转移酶 | Prupe.7G048400 | 类SAUR生长素响应蛋白 |
| SAM-dependent methyltransferases | SAUR-like auxin-responsive | ||
| Prupe.7G045400 | 吡咯啉-5-羧酸盐还原酶 | Prupe.7G048500 | 类SAUR生长素响应蛋白 |
| P5C reductase | SAUR-like auxin-responsive | ||
| Prupe.7G045500 | 吡咯啉-5-羧酸盐还原酶 | Prupe.7G048600 | 类SAUR生长素响应蛋白 |
| P5C reductase | SAUR-like auxin-responsive | ||
| Prupe.7G045600 | 未知蛋白结构247 DUF247 | Prupe.7G055900 | AGL18蛋白AGAMOUS-like 18 |
| Prupe.7G045700 | 未知蛋白结构247 | Prupe.7G056000 | 未知蛋白结构617 |
| DUF247 | Protein of unknown function,DUF617 | ||
| Prupe.7G045800 | 未知蛋白结构247 | Prupe.7G056100 | 含P环的核苷三磷酸水解酶 |
| DUF247 | P-loop containing nucleoside triphosphate hydrolases | ||
| Prupe.7G046800 | 含ankyrin重复/BTB/ POZ结构域蛋白 | Prupe.7G056200 | ABC-2型转运蛋白 |
| Ankyrin repeat family protein/BTB/POZ domain- containing protein | ABC-2 type transporter protein | ||
| Prupe.7G047000 | 甲硫氨酸依赖的甲基转移酶 | Prupe.7G056300 | 含P环的核苷三磷酸水解酶 |
| SAM-dependent methyltransferases | P-loop containing nucleoside triphosphate hydrolases | ||
| Prupe.7G047100 | PPR蛋白 | Prupe.7G056400 | ABC-2型转运蛋白 |
| Pentatricopeptide repeat protein | ABC-2 type transporter | ||
| Prupe.7G047200 | 逆转录酶 | Prupe.7G056500 | NADPH依赖的硫氧还蛋白还原酶C |
| Reverse transcriptase | NADPH-dependent thioredoxin reductase C | ||
| Prupe.7G047300 | EF-hand钙结合蛋白 | Prupe.7G056700 | 热激转录因子B3 |
| Calcium-binding EF-hand protein | Heat shock transcription factor B3 | ||
| Prupe.7G047400 | 类Patatin磷脂酶家族蛋白 | Prupe.7G056900 | SEC14细胞溶质因子/磷酸甘油酯转移酶 |
| Patatin-like phospholipase family protein | SEC14 cytosolic factor/phosphoglyceride transfer |
| [1] |
Aranzana M J, Abbassi E K, Howad W, Arús P. 2010. Genetic variation,population structure and linkage disequilibrium in peach commercial varieties. BMC Genetics, 11:69.
doi: 10.1186/1471-2156-11-69 pmid: 20646280 |
| [2] |
Bielenberg D, Wang Y, Fan S, Reighard G L, Scorza R, Abbott A G. 2004. A deletion affecting several gene candidates is present in the Evergrowing peach mutant. Journal of Heredity, 95 (5):436-444.
pmid: 15388771 |
| [3] |
Bolger A M, Lohse M, Usadel B. 2014. Trimmomatic:a flexible trimmer for Illumina sequence data. Bioinformatics, 30 (15):2114-2120.
doi: 10.1093/bioinformatics/btu170 URL |
| [4] |
Bouché N, Lauressergues D, Gasciolli V, Vaucheret H. 2006. An antagonistic function for Arabidopsis DCL2in development and a new function for DCL4 in generating viral siRNAs. The EMBO Journal, 25 (14):3347-3356.
doi: 10.1038/sj.emboj.7601217 URL |
| [5] |
Bouhadida M, Casas A M, Moreno M A, Gogorcena Y. 2007. Molecular characterization of Miraflores peach variety and relatives using SSRs. Scientia Horticulturae, 111 (2):140-145.
doi: 10.1016/j.scienta.2006.10.018 URL |
| [6] | Cai Zhi-xiang, Yan Juan, Shen Zhi-jun, Ma Rui-juan, Yu Ming-liang. 2016. Evaluation and grading of stipule length in peach germplasm resources. Journal of Plant Genetic Resources, 17 (3):71-75. (in Chinese) |
| 蔡志翔, 严娟, 沈志军, 马瑞娟, 俞明亮. 2016. 桃种质资源托叶长度评价与分级体系的建立. 植物遗传资源学报, 17 (3):71-75. | |
| [7] |
Cao K, Zheng Z, Wang L, Liu X, Zhu G, Fang W, Cheng S, Zeng P, Chen C, Wang X, Xie M, Zhong X, Wang X, Zhao P, Bian C, Zhu Y, Zhang J, Ma G, Chen C, Li Y, Hao F, Li Y, Huang G, Li Y, Li H, Guo J, Xu X, Wang J. 2014. Comparative population genomics reveals the domestication history of the peach, Prunus persica,and human influences on perennial fruit crops. Genome Biology, 15 (7):415.
doi: 10.1186/s13059-014-0415-1 pmid: 25079967 |
| [8] |
Cappa E P, El-Kassaby Y A, Garcia M N, Acu A C, Borralho N M G, Grattapaglia D, Marcucci Poltri S N, Hsiao C K. 2013. Impacts of population structure and analytical models in genome-wide association studies of complex traits in forest trees:a case study in eucalyptus globulus. PLoS ONE, 8 (11):e81267.
doi: 10.1371/journal.pone.0081267 URL |
| [9] | Chen Ji, Ma Rui-juan, Shen Zhi-jun, Cai Zhi-xiang, Yu Ming-liang. 2011. SSR analysis on the genetic relationship of ornamental peach germplasm resources. Journal of Fruit Science, 28 (4):580-585. (in Chinese) |
| 陈霁, 马瑞娟, 沈志军, 蔡志翔, 俞明亮. 2011. 基于SSR标记的观赏桃亲缘关系分析. 果树学报, 28 (4):580-585. | |
| [10] | Chen Ying-nan, Zhang Xin-ye, Dai Xiao-gang. 2014. A case study on cultivar identification in Camellia L. by using SSR markers. Nonwood Forest Research, 32 (4):140-143. (in Chinese) |
| 陈赢男, 张新叶, 戴晓港. 2014. 利用微卫星标记鉴别油茶品种. 经济林研究, 32 (4):140-143. | |
| [11] |
Cheng J, Liao L, Zhou H, Gu C, Wang L, Han Y. 2015. A small indel mutation in an anthocyanin transporter causes variegated colouration of peach flowers. Journal of Experimental Botany, 66 (22):7227-7239.
doi: 10.1093/jxb/erv419 pmid: 26357885 |
| [12] | Cheng Zhong-ping. 2003. Rapd analysis of the germplasms of Amgdalus persica var. duplex. Journal of Southwest Agricultural University(Natural science edition), 25 (1):4-6. (in Chinese) |
| 程中平. 2003. 利用RAPD技术对碧桃种质资源的分析. 西南大学学报(自然科学版), 25 (1):4-6. | |
| [13] |
de la Torre A R, Puiu D, Crepeau M W, Stevens K, Salzberg S L. 2019. Genomic architecture of complex traits in loblolly pine. New Phytologist, 221 (4):1669-1671.
doi: 10.1111/nph.2019.221.issue-4 URL |
| [14] | Dirlewanger E, Cosson P, Tavaud M, Aranzana M, Poizat C, Zanetto A, Arús P, Laigret F. 2002. Development of microsatellite markers in peach [ Prunus persica(L.)Batsch] and their use in genetic diversity analysis in peach and sweet cherry(Prunus avium L.). Theoretical & Applied Genetics, 105 (1):127-138. |
| [15] |
Du Q, Lu W, Quan M, Xiao L, Song F, Li P, Zhou D, Xie J, Wang L, Zhang D. 2018. Genome-wide association studies to improve wood properties:challenges and prospects. Frontiers in Plant Science, 9:1912.
doi: 10.3389/fpls.2018.01912 URL |
| [16] |
Dunoyer P, Himber C, Voinnet O. 2005. DICER-LIKE 4is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-tocell silencing signal. Nature Genetics, 37 (12):1356-1360.
doi: 10.1038/ng1675 URL |
| [17] | Fan Hong-hong, Xu Ting-ting, Su Jiang-shuo, Sun Wei, Zhong Xin-ran, Guan Zhi-yong, Fang Wei-min, Chen Fa-di, Zhang Fei. 2019. Identification of SNP alleles and candidate genes for cold tolerance of cut chrysanthemum. Acta Horticulturae Sinica, 46 (11):2201-2212. (in Chinese) |
| 范宏虹, 徐婷婷, 苏江硕, 孙炜, 种昕冉, 管志勇, 房伟民, 陈发棣, 张飞. 2019. 切花菊耐寒性相关SNP位点挖掘与候选基因分析. 园艺学报, 46 (11):2201-2212. | |
| [18] |
Fukudome A, Fukuhara T. 2017. Plant dicer-like proteins:double-stranded RNA-cleaving enzymes for small RNA biogenesis. Journal of Plant Research, 130 (1):33-44.
doi: 10.1007/s10265-016-0877-1 pmid: 27885504 |
| [19] | Gao Mei-ling, Zhu Zi-cheng, Gao Peng, Luan Fei-shi. 2011. A microsatellite-based genetic map of melon and localization of gene for gynoecious sex expression using recombinant inbred lines. Acta Horticulturae Sinica, 38 (7):1308-1316. (in Chinese) |
| 高美玲, 朱子成, 高鹏, 栾非时. 2011. 甜瓜重组自交系群体SSR遗传图谱构建及纯雌性基因定位. 园艺学报, 38 (7):1308-1316. | |
| [20] | Garrison E, Marth G. 2012. Haplotype-based variant detection from short-read sequencing. arXiv,1207-3907. |
| [21] |
Gasciolli V, Mallory A C, Bartel D P, Vaucheret H. 2005. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Current Biology, 15 (16):1494-1500.
pmid: 16040244 |
| [22] | He Ping. 1998. Abundance,polymorphism and applications of microsatellite in eukaryote. Hereditas, 24 (4):42-47. (in Chinese) |
| 何平. 1998. 真核生物中的微卫星及其应用. 遗传, 24 (4):42-47. | |
| [23] |
Hoshino A, Inagaki Y, Lida S. 1995. Structural analysis of Tpn1,a transposable element isolated from Japanese morning glory bearing variegated flowers. Molecular General and Genetics, 247 (1):114-117.
doi: 10.1007/BF00425828 URL |
| [24] | Li H. 2013. Aligning sequence reads,clone sequences and assembly contigs with BWA-MEM. arXiv,1303-3997. |
| [25] |
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics, 25 (16):2078-2079.
doi: 10.1093/bioinformatics/btp352 URL |
| [26] | Li Rui, Wang Tian, Wen Li-wei, Zhu Hong-liang. 2015. Research progress of Dicer-like in plants. Chinese Agricultural Sciece Bulletin, 31 (30):210-214. (in Chinese) |
| 李蕊, 王恬, 文莉薇, 朱鸿亮. 2015. 植物Dicer-like功能的研究进展. 中国农学通报, 31 (30):210-214. | |
| [27] |
Li T, Li Y, Li Z, Zhang H, Qi Y, Wang T. 2008. Simple sequence repeat analysis of genetic diversity in primary core collection of peach (Prunus persica). Journal of Integrative Plant Biology, 50 (1):102-110.
doi: 10.1111/jipb.2008.50.issue-1 URL |
| [28] |
Li Y, Wang L R. 2020. Genetic resources,breeding programs in China,and gene mining of peach:a review. Horticultural Plant Journal, 6 (4):205-215.
doi: 10.1016/j.hpj.2020.06.001 URL |
| [29] | Liu Feng, Feng Xue-mei, Zhong Wen, Liu Yu-dong, Yin Zu-jun, Han Xiu-lan, Xu Zi-chu, Shen Fa-fu. 2009. Screening of SSR core primer pairs for identificating cotton cultivar. Molecular Plant Breeding, 7 (6):1160-1168. (in Chinese) |
| 刘峰, 冯雪梅, 钟文, 刘玉栋, 阴祖军, 韩秀兰, 徐子初, 沈法富. 2009. 适合棉花品种鉴定的SSR核心引物的筛选. 分子植物育种, 7 (6):1160-1168. | |
| [30] |
Liu X J, Chuang Y N, Chiou C Y, Chin D C, Shen F Q, Yeh K W. 2012. Methylation effect on Chalcone synthase gene expression determines anthocyanin pigmentation in floral tissues of two Oncidium orchid cultivars. Planta, 236 (2):401-409.
doi: 10.1007/s00425-012-1616-z URL |
| [31] | Liu Yang, Chen Huo-ying, Wei Yu-tang. 2005. Construction of a genetic map and location ofquantitative trait loci for number of flowers per truss traits in tomato by SSR markers. Journal of Nanjing Agricultural University, 28 (4):30-34. (in Chinese) |
| 刘杨, 陈火英, 魏毓棠. 2005. 番茄SSR遗传连锁图谱的构建及每序花数性状的QTL定位. 南京农业大学学报, 28 (4):30-34. | |
| [32] |
Luan M B, Chen B F, Zou Z Z, Zhu J J, Wang X F, Xu Y, Sun Z M, Chen J H. 2015. Molecular identity of ramie germplasms using simple sequence repeat markers. Genetics and Molecular Research, 14 (1):2302-2311.
doi: 10.4238/2015.March.27.15 pmid: 25867376 |
| [33] | Luo Ran, Wu Wei-lin, Zhang Yang, Li Yu-hua. 2010. SSR marker and its application to crop genetics and breeding. Genomics and Applied Biology, 29 (1):137-143. (in Chinese) |
| 罗冉, 吴委林, 张旸, 李玉花. 2010. SSR分子标记在作物遗传育种中的应用. 基因组学与应用生物学, 29 (1):137-143. | |
| [34] | Ma Rui-juan, Zhang Yong, Yu Ming-ling, Xu Jian-lan, Shen Zhi-jun, Cai Zhi-xiang, Shen Jiang-hai. 2011. Breeding of a new peach varity Hongfenjiaren for ornamental use. Jiangsu Journal of Agricultural Science, 27 (6):1421-1422. (in Chinese) |
| 马瑞娟, 张勇, 俞明亮, 许建兰, 沈志军, 蔡志翔, 沈江海. 2011. 观赏桃新品种红粉佳人的选育. 江苏农业学报, 27 (6):1421-1422. | |
| [35] |
Mckown A D, Klapstě J, Guy R D, Geraldes A, Porth I, Hannemann J, Friedmann M, Muchero W, Tuskan G A, Ehlting J, Cronk Q C B, Elkassaby Y A, Mansfield S D, Douglas C J. 2014. Genome-wide association implicates numerous genes underlying ecological trait variation in natural populations of Populus trichocarpa. New Phytologist, 203 (2):535-553.
doi: 10.1111/nph.2014.203.issue-2 URL |
| [36] |
Mukherjee K, Campos H, Kolaczkowski B. 2013. Evolution of animal and plant dicers:early parallel duplications and recurrent adaptation of antiviral RNA binding in plants. Molecular Biology and Evolution, 30 (3):627-641.
doi: 10.1093/molbev/mss263 pmid: 23180579 |
| [37] |
Narasimhan V, Danecek P, Scally A, Xue Y, Tylersmith C, Durbin R. 2016. BCFtools/RoH:a hidden Markov model approach for detecting autozygosity from next-generation sequencing data. Bioinformatics, 32 (11):1749-1751.
doi: 10.1093/bioinformatics/btw044 pmid: 26826718 |
| [38] |
Ouni R, Zborowska A, Sehic J, Choulak S, Hormaza J I, Garkava-Gustavsson L, Mars M. 2020. Genetic diversity and structure of Tunisian local pear germplasm as revealed by SSR Markers. Horticultural Plant Journal, 6 (2):61-70.
doi: 10.1016/j.hpj.2020.03.003 URL |
| [39] |
Porth I, Klapšte J, Skyba O, Hannemann J, Mckown A D, Guy R D, Difazio S P, Muchero W, Ranjan P, Tuskan G A. 2013. Genome-wide association mapping for wood characteristics in Populus identifies an array of candidate single nucleotide polymorphisms. New Phytologist, 200 (3):710-726.
doi: 10.1111/nph.2013.200.issue-3 URL |
| [40] |
Purcell S, Neale B M, Toddbrown K, Thomas L, Ferreira M A R, Bender D, Maller J, Sklar P, De Bakker P I W, Daly M J. 2007. PLINK:A tool set for whole-genome association and population-based linkage analyses. The American Journal of Human Genetics, 81 (3):559-575.
doi: 10.1086/519795 URL |
| [41] |
Quattrocchio F, Wing J F, Der Woude K V, Souer E, De Vetten N, Mol J N M, Koes R. 1999. Molecular analysis of the anthocyanin2gene of petunia and its role in the evolution of flower color. The Plant Cell, 11 (8):1433-1444.
doi: 10.1105/tpc.11.8.1433 URL |
| [42] |
Schauer S E, Jacobsen S E, Meinke D W, Ray A. 2002. DICER-LIKE 1:blind men and elephants in Arabidopsis development. Trends in Plant Science, 7 (11):487-491.
doi: 10.1016/S1360-1385(02)02355-5 URL |
| [43] |
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6:molecular evolutionary genetics analysis version 6.0. Molecular Biology Evolution, 30 (12):2725-2729.
doi: 10.1093/molbev/mst197 URL |
| [44] | Wang Haoying, Liu Zhiyuan, Wang Xiaowu, Wu Jian, Zhang Helong, Xia Zhilan, Xu Zhaosheng, Qian Wei. 2019. Genome-wide association studies for monoecism in spinach. Acta Horticulturae Sinica, 46 (8):1495-1502. (in Chinese) |
| 汪豪英, 刘志远, 王晓武, 武剑, 张合龙, 夏志兰, 徐兆生, 钱伟. 2019. 菠菜雌雄同株性状的全基因组关联分析. 园艺学报, 46 (8):1495-1502. | |
| [45] |
Wang Z, Meng D, Wang A, Li T, Jiang S, Li C T. 2013. The methylation of the PcMYB10 promoter is ssociated with green-skinned sport in max red bartlett pear. Plant Physiology, 162 (2):885-896.
doi: 10.1104/pp.113.214700 URL |
| [46] | Wang Zu-hua, Zhou Jian-tao. 1990. Pollen morphology of peach germplasm. Acta Horticulturae Sinica, 17 (3):161-168. (in Chinese) |
| 汪祖华, 周建涛. 1990. 桃种质的亲缘演化关系研究——花粉形态分析. 园艺学报, 17 (3):161-168. | |
| [47] |
Wigginton J E, Cutler D J, Abecasis G R. 2005. A note on exact tests of hardy-weinberg equilibrium. The American Journal of Human Genetics, 76 (5):887-893.
doi: 10.1086/429864 URL |
| [48] | Wu Q, Liang X, Dai X, Chen Y, Yin T. 2018. Molecular discrimination and ploidy level determination for elite willow cultivars. Tree Genetics & Genomes, 14 (5):65. |
| [49] | Wu X, Zhou Y, Yao D, Iqbal S, Zhang Z. 2020. DNA methylation of LDOX gene contributes to the floral colour variegation in peach. Journal of Plant Physiology,246-247:153116. |
| [50] | Wünsch A, Carrera M, Hormaza J I. 2006. Molecular characterization of local spanish peach [ Prunus persica (L.)Batsch] germplasm. Genetic Resources & Crop Evolution, 53 (5):925-932. |
| [51] | Xiang Yuanping, Wang Yidan, He Hongjun, Xu Qijiang. 2020. Plant transposable elements and the effects of insertion mutations on flower development in horticultural plants. Acta Horticulturae Sinica, 47 (11):2247-2266. (in Chinese) |
| 相元萍, 王一丹, 贺洪军, 徐启江. 2020. 植物转座子类型及其插入突变对园艺植物花发育影响的研究进展. 园艺学报, 47 (11):2247-2266. | |
| [52] |
Xie Z, Johansen L K, Gustafson A M, Kasschau K D, Lellis A D, Zilberman D, Jacobsen S E, Carrington J C. 2004. Genetic and functional diversification of small RNA pathways in plants. PLoS Biology, 2 (5):e104.
doi: 10.1371/journal.pbio.0020104 URL |
| [53] |
Xu M, Brar H K, Grosic S, Palmer R G, Bhattacharyya M K. 2010. Excision of an active CACTA-like transposable element from DFR2 causes variegated flowers in soybean[ Glycine max(L.) Merr.]. Genetics, 184 (1):53-63.
doi: 10.1534/genetics.109.107904 |
| [54] |
Yan F, Di S, Rojas Rodas F, Rodriguez Torrico T, Murai Y, Iwashina T, Anai T, Takahashi R. 2014. Allelic variation of soybean flower color gene W4 encoding dihydroflavonol 4-reductase 2. BMC Plant Biology, 14:58.
doi: 10.1186/1471-2229-14-58 URL |
| [55] |
Yoshikawa M, Peragine A, Park M Y, Poethig R S. 2005. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes development, 19 (18):2164-2175.
doi: 10.1101/gad.1352605 URL |
| [56] |
Zhang H, Lang Z, Zhu J K. 2018. Dynamics and function of DNA methylation in plants. Nature Reviews Molecular Cell Biology, 19 (8):489-506.
doi: 10.1038/s41580-018-0016-z URL |
| [57] | Zhang Yan-ming, Xing Guo-fang, Liu Mei-tao, Liu Xiao-dong, Han Yuan-huai. 2013. Genome wide association study:opportunities and challenges in genomic research. Biotechnology Bulletin,(6):1-6. (in Chinese) |
| 张雁明, 邢国芳, 刘美桃, 刘晓东, 韩渊怀. 2013. 全基因组关联分析:基因组学研究的机遇与挑战. 生物技术通报,(6):1-6. | |
| [58] |
Zhou X, Stephens M. 2012. Genome-wide efficient mixed model analysis for association studies. Nature Genetics, 44 (7):821-824.
doi: 10.1038/ng.2310 URL |
| [59] | Zhu Geng-rui, Wang Li-rong, Fang Wei-chao, Cao Ke, Chen Chang-wen, Li Quan-hong, Feng Yi-bin, Ling Guo-jun, Yue Zhang-ping. 2011. A low chilling and early blooming ornamental peach cultivar‘Baochun’. Acta Horticulturae Sinica, 38 (10):2035-2036. (in Chinese) |
| 朱更瑞, 王力荣, 方伟超, 曹珂, 陈昌文, 李全红, 冯义彬, 凌国钧, 岳长平. 2011. 低需冷量早花观赏桃品种‘报春’. 园艺学报, 38 (10):2035-2036. |
| [1] | 乔军, 刘婧, 李素文, 王利英. 基于极端混合池全基因组重测序的茄子萼下果色基因预测[J]. 园艺学报, 2022, 49(3): 613-621. |
| [2] | 卢甜甜, 刘志远, 徐兆生, 张合龙, 李国亮, 折红兵, 钱伟. 菜豆花色全基因组关联分析[J]. 园艺学报, 2022, 49(2): 332-340. |
| [3] | 刘根忠, 石春美, 于会洋, 王莹, 陈卫芳, 尚乐乐, 张余洋, 叶志彪. 番茄节间长度基因IL10和IL11的鉴定和功能验证[J]. 园艺学报, 2021, 48(7): 1340-1348. |
| [4] | 贾会霞, 李锡香, 宋江萍, 林毓娥, 张晓辉, 邱杨, 阳文龙, 娄群峰, 王海平. 黄瓜核心种质白粉病抗性的全基因组关联分析[J]. 园艺学报, 2021, 48(7): 1371-1385. |
| [5] | 江锡兵, 章平生, 徐阳, 吴聪连, 张东北, 龚榜初, 吴开云, 赖俊声. 栗杂交F1代SSR标记遗传多样性分析[J]. 园艺学报, 2021, 48(5): 897-907. |
| [6] | 董艺, 冯羽飞, 许忠民, 王世民, 唐鸿吕, 黄炜. SSR标记遗传距离与结球甘蓝杂种优势的关系分析[J]. 园艺学报, 2021, 48(5): 934-946. |
| [7] | 蒋爽, 张学英, 安海山, 徐芳杰, 章加应. 枇杷全基因组SSR标记开发及其多态性研究[J]. 园艺学报, 2021, 48(5): 1013-1022. |
| [8] | 范宏虹,徐婷婷,苏江硕,孙 炜,种昕冉,管志勇,房伟民,陈发棣,张 飞*. 切花菊耐寒性相关SNP位点挖掘与候选基因分析[J]. 园艺学报, 2019, 46(11): 2201-2212. |
| [9] | 夏岩石1,张润霖1,卢宇鹏1,李荣华1,李光光2,张 华2,郭培国1,*. 菜薹品质性状与SSR标记的关联分析[J]. 园艺学报, 2019, 46(10): 1960-1972. |
| [10] | 刘 磊,邓学斌,冯晶晶,王 静,孙晓荣,舒金帅,李君明*. 加工番茄Ve-1、I-2、Mi-1和Pto基因关联变异位点的挖掘[J]. 园艺学报, 2018, 45(6): 1089-1100. |
| [11] | 袁王俊1,叶 松2,董美芳2,韩远记2,张维瑞1,尚富德2,*. 菊花品种表型性状与SSR和SCoT分子标记的关联分析[J]. 园艺学报, 2017, 44(2): 364-372. |
| [12] | 周宁宁,李淑斌,李远波,蹇洪英,晏慧君,王其刚,陈 敏,邱显钦,张 颢,唐开学*. 二倍体月季F1群体的SSR鉴定与遗传分析[J]. 园艺学报, 2017, 44(1): 151-160. |
| [13] | 孙玉燕, 邱杨, 段蒙蒙, 陈晓华, 段韫丹, 吕美洁, 杨好慧, 李锡香. 萝卜叶片干枯基因的定位[J]. 园艺学报, 2015, 42(8): 1505-1514. |
| [14] | 杨树华1,李世超1,贾瑞冬1,赵 鑫1,金茂勇2,李凤杰2,葛 红1,*. 金叶弯刺蔷薇× 山刺玫遗传连锁图谱的初步构建[J]. 园艺学报, 2015, 42(12): 2526-2534. |
| [15] | 刘源霞1,2,李保华3,王彩虹2,刘春晓2,孔祥华2,祝 军2,戴洪义2,*. 苹果对炭疽菌叶枯病抗性遗传的研究及其分子标记筛选[J]. 园艺学报, 2015, 42(11): 2105-2112. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司